|
Size: 45735
Comment:
|
Size: 62750
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 6: | Line 6: |
| Line 9: | Line 10: |
| {{{ | {{{#!sagecell |
| Line 28: | Line 29: |
| html("<h1>Double Precision Root Finding Using Bisection</h1>") @interact def _(f = cos(x) - x, a = float(0), b = float(1), eps=(-3,(-16..-1))): |
pretty_print(html("<h1>Double Precision Root Finding Using Bisection</h1>")) @interact def _(f = cos(x) - x, a = float(0), b = float(1), eps=(-3,[-16..-1])): |
| Line 34: | Line 35: |
| time c, intervals = bisect_method(f, a, b, eps) | c, intervals = bisect_method(f, a, b, eps) |
| Line 56: | Line 57: |
| {{{ | https://cloud.sagemath.com/projects/19575ea0-317e-402b-be57-368d04c113db/files/pub/2801-2901/2824-Double%20Precision%20Root%20Finding%20Using%20Newton's%20Method.sagews {{{#!sagecell |
| Line 72: | Line 75: |
| html("<h1>Double Precision Root Finding Using Newton's Method</h1>") | var('x') pretty_print(html("<h1>Double Precision Root Finding Using Newton's Method</h1>")) |
| Line 77: | Line 82: |
| time z, iterates = newton_method(f, c, eps) | z, iterates = newton_method(f, c, eps) |
| Line 79: | Line 84: |
| print "f(c) = %r"%f(z) | print "f(c) = %r"%f(x=z) |
| Line 82: | Line 87: |
| html(iterates) P = plot(f, z-interval, z+interval, rgbcolor='blue') |
pretty_print(html(iterates)) P = plot(f, (x,z-interval, z+interval), rgbcolor='blue') |
| Line 94: | Line 99: |
| http://sagenb.org/home/pub/2823/ {{{ |
https://cloud.sagemath.com/projects/19575ea0-317e-402b-be57-368d04c113db/files/pub/2801-2901/2823.sagews {{{#!sagecell |
| Line 112: | Line 117: |
| {{{ html('<h2>Tangent line grapher</h2>') |
{{{#!sagecell pretty_print(html('<h2>Tangent line grapher</h2>')) |
| Line 124: | Line 129: |
| fmax = f.find_maximum_on_interval(prange[0], prange[1])[0] fmin = f.find_minimum_on_interval(prange[0], prange[1])[0] |
fmax = f.find_local_maximum(prange[0], prange[1])[0] fmin = f.find_local_minimum(prange[0], prange[1])[0] |
| Line 132: | Line 137: |
| {{{ | {{{#!sagecell #find_maximum_on_interval and find_minimum_on_interval are deprecated #use find_local_maximum find_local_minimum instead #see http://trac.sagemath.org/2607 for details -RRubalcaba |
| Line 147: | Line 156: |
| min_y = find_minimum_on_interval(func,a,b)[0] max_y = find_maximum_on_interval(func,a,b)[0] html('<h3>Numerical integrals with the midpoint rule</h3>') html('$\int_{a}^{b}{f(x) dx} {\\approx} \sum_i{f(x_i) \Delta x}$') |
min_y = min(0, sage.numerical.optimize.find_local_minimum(func,a,b)[0]) max_y = max(0, sage.numerical.optimize.find_local_maximum(func,a,b)[0]) pretty_print(html('<h3>Numerical integrals with the midpoint rule</h3>')) pretty_print(html('$\int_{a}^{b}{f(x) dx} {\\approx} \sum_i{f(x_i) \Delta x}$')) |
| Line 160: | Line 169: |
| {{{ | {{{#!sagecell |
| Line 162: | Line 171: |
| #find_maximum_on_interval and find_minimum_on_interval are deprecated #use find_local_maximum find_local_minimum instead #see http://trac.sagemath.org/2607 for details -RRubalcaba |
|
| Line 185: | Line 197: |
| x = find_maximum_on_interval(func, q*dx + a, q*dx + dx + a)[1] | x = find_local_maximum(func, q*dx + a, q*dx + dx + a)[1] |
| Line 188: | Line 200: |
| x = find_minimum_on_interval(func, q*dx + a, q*dx + dx + a)[1] | x = find_local_minimum(func, q*dx + a, q*dx + dx + a)[1] |
| Line 199: | Line 211: |
| min_y = min(0, find_minimum_on_interval(func,a,b)[0]) max_y = max(0, find_maximum_on_interval(func,a,b)[0]) |
min_y = min(0, find_local_minimum(func,a,b)[0]) max_y = max(0, find_local_maximum(func,a,b)[0]) |
| Line 216: | Line 228: |
| html(r''' | pretty_print(html(r''' |
| Line 226: | Line 238: |
| ''' % (numerical_answer, number_of_subdivisions, sum_html, num_html, estimated_answer)) | ''' % (numerical_answer, number_of_subdivisions, sum_html, num_html, estimated_answer))) |
| Line 233: | Line 245: |
| {{{ | {{{#!sagecell |
| Line 246: | Line 258: |
| {{{ | {{{#!sagecell |
| Line 344: | Line 356: |
| {{{ | {{{#!sagecell |
| Line 446: | Line 458: |
| == Coordinate Transformations == | == Coordinate Transformations (FIXME in Jupyter) == |
| Line 450: | Line 462: |
| {{{ | {{{#!sagecell |
| Line 452: | Line 464: |
| # polar coordinates #(x,y)=(u*cos(v),u*sin(v)); (u_range,v_range)=([0..6],[0..2*pi,step=pi/12]) # weird example (x,y)=(u^2-v^2,u*v+cos(u*v)); (u_range,v_range)=([-5..5],[-5..5]) thickness=4 square_length=.05 |
|
| Line 455: | Line 476: |
| def trans(x=input_box(u^2-v^2, label="x=",type=SR), \ y=input_box(u*v+cos(u*v), label="y=",type=SR), \ t_val=slider(0,10,0.2,6, label="Length of curves"), \ |
def trans(x=input_box(x, label="x",type=SR), y=input_box(y, label="y",type=SR), |
| Line 460: | Line 480: |
| u_range=input_box(range(-5,5,1), label="u lines"), v_range=input_box(range(-5,5,1), label="v lines")): thickness=4 u_val = min(u_range)+(max(u_range)-min(u_range))*u_percent v_val = min(v_range)+(max(v_range)-min(v_range))*v_percent t_min = -t_val t_max = t_val g1=sum([parametric_plot((i,v), (v,t_min,t_max), rgbcolor=(1,0,0)) for i in u_range]) g2=sum([parametric_plot((u,i), (u,t_min,t_max), rgbcolor=(0,0,1)) for i in v_range]) vline_straight=parametric_plot((u,v_val), (u,t_min,t_max), rgbcolor=(0,0,1), linestyle='-',thickness=thickness) uline_straight=parametric_plot((u_val, v), (v,t_min,t_max),rgbcolor=(1,0,0), linestyle='-',thickness=thickness) (g1+g2+vline_straight+uline_straight).save("uv_coord.png",aspect_ratio=1, figsize=[5,5], axes_labels=['$u$','$v$']) xuv = fast_float(x,'u','v') yuv = fast_float(y,'u','v') xvu = fast_float(x,'v','u') yvu = fast_float(y,'v','u') g3=sum([parametric_plot((partial(xuv,i),partial(yuv,i)), (v,t_min,t_max), rgbcolor=(1,0,0)) for i in u_range]) g4=sum([parametric_plot((partial(xvu,i),partial(yvu,i)), (u,t_min,t_max), rgbcolor=(0,0,1)) for i in v_range]) uline=parametric_plot((partial(xuv,u_val),partial(yuv,u_val)),(v,t_min,t_max),rgbcolor=(1,0,0), linestyle='-',thickness=thickness) vline=parametric_plot((partial(xvu,v_val),partial(yvu,v_val)), (u,t_min,t_max), rgbcolor=(0,0,1), linestyle='-',thickness=thickness) (g3+g4+vline+uline).save("xy_coord.png", aspect_ratio=1, figsize=[5,5], axes_labels=['$x$','$y$']) print jsmath("x=%s, \: y=%s"%(latex(x), latex(y))) print "<html><table><tr><td><img src='cell://uv_coord.png'/></td><td><img src='cell://xy_coord.png'/></td></tr></table></html>" |
t_val=slider(0,10,0.2,6, label="Length"), u_range=input_box(u_range, label="u lines"), v_range=input_box(v_range, label="v lines")): x(u,v)=x y(u,v)=y u_val = min(u_range)+(max(u_range)-min(u_range))*u_percent v_val = min(v_range)+(max(v_range)-min(v_range))*v_percent t_min = -t_val t_max = t_val uvplot=sum([parametric_plot((i,v), (v,t_min,t_max), color='red',axes_labels=['u','v'],figsize=[5,5]) for i in u_range]) uvplot+=sum([parametric_plot((u,i), (u,t_min,t_max), color='blue',axes_labels=['u','v']) for i in v_range]) uvplot+=parametric_plot((u,v_val), (u,t_min,t_max), rgbcolor=(0,0,1), linestyle='-',thickness=thickness) uvplot+=parametric_plot((u_val, v), (v,t_min,t_max),rgbcolor=(1,0,0), linestyle='-',thickness=thickness) pt=vector([u_val,v_val]) du=vector([(t_max-t_min)*square_length,0]) dv=vector([0,(t_max-t_min)*square_length]) uvplot+=polygon([pt,pt+dv,pt+du+dv,pt+du],color='purple',alpha=0.7) uvplot+=line([pt,pt+dv,pt+du+dv,pt+du],color='green') T(u,v)=(x,y) xuv = fast_float(x,'u','v') yuv = fast_float(y,'u','v') xvu = fast_float(x,'v','u') yvu = fast_float(y,'v','u') xyplot=sum([parametric_plot((partial(xuv,i),partial(yuv,i)), (v,t_min,t_max), color='red', axes_labels=['x','y'],figsize=[5,5]) for i in u_range]) xyplot+=sum([parametric_plot((partial(xvu,i),partial(yvu,i)), (u,t_min,t_max), color='blue') for i in v_range]) xyplot+=parametric_plot((partial(xuv,u_val),partial(yuv,u_val)),(v,t_min,t_max),color='red', linestyle='-',thickness=thickness) xyplot+=parametric_plot((partial(xvu,v_val),partial(yvu,v_val)), (u,t_min,t_max), color='blue', linestyle='-',thickness=thickness) jacobian=abs(T.diff().det()).simplify_full() t_vals=[0..1,step=t_val*.01] vertices=[(x(*c),y(*c)) for c in [pt+t*dv for t in t_vals]] vertices+=[(x(*c),y(*c)) for c in [pt+dv+t*du for t in t_vals]] vertices+=[(x(*c),y(*c)) for c in [pt+(1-t)*dv+du for t in t_vals]] vertices+=[(x(*c),y(*c)) for c in [pt+(1-t)*du for t in t_vals]] xyplot+=polygon(vertices,color='purple',alpha=0.7) xyplot+=line(vertices,color='green') html("$T(u,v)=%s$"%(latex(T(u,v)))) html("Jacobian: $%s$"%latex(jacobian(u,v))) html("A very small region in $xy$ plane is approximately %0.4g times the size of the corresponding region in the $uv$ plane"%jacobian(u_val,v_val).n()) pretty_print(table([[uvplot,xyplot]])) |
| Line 492: | Line 529: |
| {{{ | {{{#!sagecell |
| Line 515: | Line 552: |
| {{{ | {{{#!sagecell |
| Line 542: | Line 579: |
| {{{ | {{{#!sagecell |
| Line 568: | Line 605: |
| {{{ | {{{#!sagecell |
| Line 583: | Line 620: |
| {{{ | {{{#!sagecell |
| Line 602: | Line 639: |
| sin,cos = math.sin,math.cos | |
| Line 629: | Line 666: |
| {{{ from scipy.special.orthogonal import p_roots |
{{{#!sagecell import scipy import numpy from scipy.special.orthogonal import p_roots, t_roots, u_roots |
| Line 639: | Line 678: |
| 'Chebyshev': {'w': 1/sqrt(1-x**2), 'xmin': -1, 'xmax': 1, 'func': t_roots}, 'Chebyshev2': {'w': sqrt(1-x**2), 'xmin': -1, 'xmax': 1, 'func': u_roots}, 'Trapezoid': {'w': 1, 'xmin': -1, 'xmax': 1, 'func': lambda n: (linspace(-1r,1,n), numpy.array([1.0r]+[2.0r]*(n-2)+[1.0r])*1.0r/n)}, 'Simpson': {'w': 1, 'xmin': -1, 'xmax': 1, 'func': lambda n: (linspace(-1r,1,n), numpy.array([1.0r]+[4.0r,2.0r]*int((n-3.0r)/2.0r)+[4.0r,1.0r])*2.0r/(3.0r*n))}} |
'Chebyshev': {'w': 1/sqrt(1-x**2), 'xmin': -1, 'xmax': 1, 'func': t_roots}, 'Chebyshev2': {'w': sqrt(1-x**2), 'xmin': -1, 'xmax': 1, 'func': u_roots}, 'Trapezoid': {'w': 1, 'xmin': -1, 'xmax': 1, 'func': lambda n: (linspace(-1r,1,n), numpy.array([1.0r]+[2.0r]*(n-2)+[1.0r])*1.0r/n)}, 'Simpson': {'w': 1, 'xmin': -1, 'xmax': 1, 'func': lambda n: (linspace(-1r,1,n), numpy.array([1.0r]+[4.0r,2.0r]*int((n-3.0r)/2.0r)+[4.0r,1.0r])*2.0r/(3.0r*n))}} |
| Line 646: | Line 688: |
| return polygon([(center-width2,0),(center+width2,0),(center+width2,height),(center-width2,height)],**kwds) | return polygon([(center-width2,0), (center+width2,0),(center+width2,height),(center-width2,height)],**kwds) |
| Line 650: | Line 693: |
| def weights(n=slider(1,30,1,default=10),f=input_box(default=3*x+cos(10*x)),show_method=["Legendre", "Chebyshev", "Chebyshev2", "Trapezoid","Simpson"]): | def weights(n=slider(1,30,1,default=10),f=input_box(default=3*x+cos(10*x),type=SR), show_method=["Legendre", "Chebyshev", "Chebyshev2", "Trapezoid","Simpson"]): |
| Line 659: | Line 703: |
| scaled_ff = fast_float(scaled_func) | scaled_ff = fast_float(scaled_func, 'x') |
| Line 667: | Line 711: |
| stems = sum(line([(x,0),(x,scaled_ff(x))],rgbcolor=(1-y,1-y,1-y),thickness=2,markersize=6,alpha=y) for x,y in coords_scaled) points = sum([point([(x,0),(x,scaled_ff(x))],rgbcolor='black',pointsize=30) for x,_ in coords]) |
stems = sum(line([(x,0),(x,scaled_ff(x))],rgbcolor=(1-y,1-y,1-y), thickness=2,markersize=6,alpha=y) for x,y in coords_scaled) points = sum([point([(x,0), (x,scaled_ff(x))],rgbcolor='black',pointsize=30) for x,_ in coords]) |
| Line 673: | Line 719: |
| show(graph,xmin=plot_min,xmax=plot_max) | show(graph,xmin=plot_min,xmax=plot_max,aspect_ratio="auto") |
| Line 681: | Line 727: |
| html("$$\sum_{i=1}^{i=%s}w_i\left(%s\\right)= %s\\approx %s =\int_{-1}^{1}%s \,dx$$"%(n,latex(f.subs(x="x_i")), approximation, integral, latex(scaled_func))) | html("$$\sum_{i=1}^{i=%s}w_i\left(%s\\right)= %s\\approx %s =\int_{-1}^{1}%s \,dx$$"%(n, latex(f), approximation, integral, latex(scaled_func))) |
| Line 694: | Line 741: |
| {{{ | {{{#!sagecell |
| Line 725: | Line 772: |
| velocity = derivative( position(t) ) acceleration = derivative(velocity(t)) |
velocity = derivative(position(t), t) acceleration = derivative(velocity(t), t) |
| Line 728: | Line 775: |
| speed_deriv = derivative(speed) | speed_deriv = derivative(speed, t) |
| Line 730: | Line 777: |
| dT = derivative(tangent(t)) | dT = derivative(tangent(t), t) |
| Line 801: | Line 848: |
| == Vector Calculus, 3-D Motion == | == Vector Calculus, 3-D Motion (FIXME) == |
| Line 806: | Line 853: |
| {{{ | {{{#!sagecell |
| Line 843: | Line 890: |
| velocity = derivative( position(t) ) acceleration = derivative(velocity(t)) |
velocity = derivative( position(t), t) acceleration = derivative(velocity(t), t) |
| Line 846: | Line 893: |
| speed_deriv = derivative(speed) | speed_deriv = derivative(speed, t) |
| Line 848: | Line 895: |
| dT = derivative(tangent(t)) | dT = derivative(tangent(t), t) |
| Line 851: | Line 898: |
| ## dB = derivative(binormal(t)) | ## dB = derivative(binormal(t), t) |
| Line 929: | Line 976: |
| == Multivariate Limits by Definition == by John Travis http://sagenb.mc.edu/home/pub/97/ {{{#!sagecell ## An interactive way to demonstrate limits of multivariate functions of the form z = f(x,y) ## ## John Travis ## Mississippi College ## ## Spring 2011 ## # Starting point for radius values before collapsing in as R approaches 0. # Functions ought to be "reasonable" within a circular domain of radius R surrounding # the desired (x_0,y_0). var('x,y,z') Rmin=1/10 Rmax=2 @interact(layout=dict(top=[['f'],['x0'],['y0']], bottom=[['in_3d','curves','R','graphjmol']])) def _(f=input_box((x^2-y^2)/(x^2+y^2),width=30,label='$f(x)$'), R=slider(Rmin,Rmax,1/10,Rmax,label=', $R$'), x0=input_box(0,width=10,label='$x_0$'), y0=input_box(0,width=10,label='$y_0$'), curves=checkbox(default=false,label='Show curves'), in_3d=checkbox(default=false,label='3D'), graphjmol=checkbox(default=true,label='Interactive graph')): if graphjmol: view_method = 'jmol' else: view_method = 'tachyon' # converting f to cylindrical coordinates. g(r,t) = f(x=r*cos(t)+x0,y=r*sin(t)+y0) # Sage graphing transformation used to see the original surface. cylinder = (r*cos(t)+x0,r*sin(t)+y0,z) surface = plot3d(g,(t,0,2*pi),(r,1/100,Rmax),transformation=cylinder,opacity=0.2) # Regraph the surface for smaller and smaller radii controlled by the slider. collapsing_surface = plot3d(g,(t,0,2*pi),(r,1/100,R),transformation=cylinder,rgbcolor=(0,1,0)) G = surface+collapsing_surface html('Enter $(x_0 ,y_0 )$ above and see what happens as $ R \\rightarrow 0 $.') html('The surface has a limit as $(x,y) \\rightarrow $ ('+str(x0)+','+str(y0)+') if the green region collapses to a point.') # If checked, add a couple of curves on the surface corresponding to limit as x->x0 for y=x^(3/5), # and as y->y0 for x=y^(3/5). Should make this more robust but perhaps using # these relatively obtuse curves could eliminate problems. if curves: curve_x = parametric_plot3d([x0-t,y0-t^(3/5),f(x=x0-t,y=y0-t^(3/5))],(t,Rmin,Rmax),color='red',thickness=10) curve_y = parametric_plot3d([x0+t^(3/5),y0+t,f(x=x0+t^(3/5),y=y0+t)],(t,Rmin,Rmax),color='red',thickness=10) R2 = Rmin/4 G += arrow((x0-Rmin,y0-Rmin^(3/5),f(x=x0-Rmin,y=y0-Rmin^(3/5))),(x0-R2,y0-R2^(3/5),f(x=x0-R2,y=y0-R2^(3/5))),size=30 ) G += arrow((x0+Rmin^(3/5),y0+Rmin,f(x=x0+Rmin^(3/5),y=y0+Rmin)),(x0+R2^(3/5),y0+R2,f(x=x0+R2^(3/5),y=y0+R2)),size=30 ) limit_x = limit(f(x=x0-t,y=y0-t^(3/5)),t=0) limit_y = limit(f(x=x0+t^(3/5),y=y0+t),t=0) text_x = text3d(limit_x,(x0,y0,limit_x)) text_y = text3d(limit_y,(x0,y0,limit_y)) G += curve_x+curve_y+text_x+text_y html('The red curves represent a couple of trajectories on the surface. If they do not meet, then') html('there is also no limit. (If computer hangs up, likely the computer can not do these limits.)') html('\n<center><font color="red">$\lim_{(x,?)\\rightarrow(x_0,y_0)} f(x,y) =%s$</font>'%str(limit_x)+' and <font color="red">$\lim_{(?,y)\\rightarrow(x_0,y_0)} f(x,y) =%s$</font></center>'%str(limit_y)) if in_3d: show(G,stereo="redcyan",viewer=view_method) else: show(G,perspective_depth=true,viewer=view_method) }}} {{attachment:3D_Limit_Defn.png}} {{{#!sagecell ## An interactive way to demonstrate limits of multivariate functions of the form z = f(x,y) ## This one uses contour plots and so will work with functions that have asymptotic behavior. ## ## John Travis ## Mississippi College ## ## Spring 2011 ## # An increasing number of contours for z = f(x,y) are utilized surrounding a desired (x_0,y_0). # A limit can be shown to exist at (x_0,y_0) provided the point stays trapped between adjacent # contour lines as the number of lines increases. If the contours change wildly near the point, # then a limit does not exist. # Looking for two different paths to approach (x_0,y_0) that utilize a different selection of colors # will help locate paths to use that exhibit the absence of a limit. var('x,y,z,u') @interact(layout=dict(top=[['f'],['x0'],['y0']], bottom=[['N'],['R']])) def _(f=input_box(default=(x*y^2)/(x^2+y^4),width=30,label='$f(x)$'), N=slider(5,100,1,10,label='Number of Contours'), R=slider(0.1,1,0.01,1,label='Radius of circular neighborhood'), x0=input_box(0,width=10,label='$x_0$'), y0=input_box(0,width=10,label='$y_0$')): html('Enter $(x_0 ,y_0 )$ above and see what happens as the number of contour levels $\\rightarrow \infty $.') html('A surface will have a limit in the center of this graph provided there is not a sudden change in color there.') # Need to make certain the min and max contour lines are not huge due to asymptotes. If so, clip and start contours at some reasonable # values so that there are a nice collection of contours to show around the desired point. surface = contour_plot(f,(x,x0-1,x0+1),(y,y0-1,y0+1),cmap=True,colorbar=True,fill=False,contours=N) surface += parametric_plot([R*cos(u),R*sin(u)],[0,2*pi],color='black') # Nice to use if f=x*y^2/(x^2 + y^4) # var('u') # surface += parametric_plot([u^2,u],[u,-1,1],color='black') limit_point = point((x0,y0),color='red',size=30) # show(limit_point+surface) pretty_print(table([[surface],['hi']])) }}} {{attachment:3D_Limit_Defn_Contours.png}} |
|
| Line 933: | Line 1104: |
| {{{ | {{{#!sagecell |
| Line 986: | Line 1157: |
| {{{ %hide %auto |
{{{#!sagecell |
| Line 1047: | Line 1216: |
| {{{ | {{{#!sagecell |
| Line 1085: | Line 1254: |
| {{{ | {{{#!sagecell |
| Line 1118: | Line 1287: |
| == Parametric surface example == | == Volumes over non-rectangular domains == by John Travis https://cloud.sagemath.com/projects/19575ea0-317e-402b-be57-368d04c113db/files/pub/2801-2901/2829.sagews {{{#!sagecell ## Graphing surfaces over non-rectangular domains ## John Travis ## Spring 2011 ## ## ## An updated version of this worksheet may be available at http://sagenb.mc.edu ## ## Interact allows the user to input up to two inequality constraints on the ## domain when dealing with functional surfaces ## ## User inputs: ## f = "top" surface with z = f(x,y) ## g = "bottom" surface with z = g(x,y) ## condition1 = a single boundary constraint. It should not include && or | to join two conditions. ## condition2 = another boundary constraint. If there is only one constraint, just enter something true ## or even just an x (or y) in the entry blank. ## ## var('x,y') # f is the top surface # g is the bottom surface global f,g # condition1 and condition2 are the inequality constraints. It would be nice # to have any number of conditions connected by $$ or | global condition1,condition2 @interact def _(f=input_box(default=(1/3)*x^2 + (1/4)*y^2 + 5,label='$f(x)=$'), g=input_box(default=-1*x+0*y,label='$g(x)=$'), condition1=input_box(default= x^2+y^2<8,label='$Constraint_1=$'), condition2=input_box(default=y<sin(3*x),label='$Constraint_2=$'), show_3d=('Stereographic',false), show_vol=('Shade volume',true), dospin = ('Spin?',true), clr = color_selector('#faff00', label='Volume Color', widget='colorpicker', hide_box=True), xx = range_slider(-5, 5, 1, default=(-3,3), label='X Range'), yy = range_slider(-5, 5, 1, default=(-3,3), label='Y Range'), auto_update=false): # This is the top function actually graphed by using NaN outside domain def F(x,y): if condition1(x=x,y=y): if condition2(x=x,y=y): return f(x=x,y=y) else: return -NaN else: return -NaN # This is the bottom function actually graphed by using NaN outside domain def G(x,y): if condition1(x=x,y=y): if condition2(x=x,y=y): return g(x=x,y=y) else: return -NaN else: return -NaN P = Graphics() # The graph of the top and bottom surfaces P_list = [] P_list.append(plot3d(F,(x,xx[0],xx[1]),(y,yy[0],yy[1]),color='blue',opacity=0.9)) P_list.append(plot3d(G,(x,xx[0],xx[1]),(y,yy[0],yy[1]),color='gray',opacity=0.9)) # Interpolate "layers" between the top and bottom if desired if show_vol: ratios = range(10) def H(x,y,r): return (1-r)*F(x=x,y=y)+r*G(x=x,y=y) P_list.extend([ plot3d(lambda x,y: H(x,y,ratios[1]/10),(x,xx[0],xx[1]),(y,yy[0],yy[1]),opacity=0.2,color=clr), plot3d(lambda x,y: H(x,y,ratios[2]/10),(x,xx[0],xx[1]),(y,yy[0],yy[1]),opacity=0.2,color=clr), plot3d(lambda x,y: H(x,y,ratios[3]/10),(x,xx[0],xx[1]),(y,yy[0],yy[1]),opacity=0.2,color=clr), plot3d(lambda x,y: H(x,y,ratios[4]/10),(x,xx[0],xx[1]),(y,yy[0],yy[1]),opacity=0.2,color=clr), plot3d(lambda x,y: H(x,y,ratios[5]/10),(x,xx[0],xx[1]),(y,yy[0],yy[1]),opacity=0.2,color=clr), plot3d(lambda x,y: H(x,y,ratios[6]/10),(x,xx[0],xx[1]),(y,yy[0],yy[1]),opacity=0.2,color=clr), plot3d(lambda x,y: H(x,y,ratios[7]/10),(x,xx[0],xx[1]),(y,yy[0],yy[1]),opacity=0.2,color=clr), plot3d(lambda x,y: H(x,y,ratios[8]/10),(x,xx[0],xx[1]),(y,yy[0],yy[1]),opacity=0.2,color=clr), plot3d(lambda x,y: H(x,y,ratios[9]/10),(x,xx[0],xx[1]),(y,yy[0],yy[1]),opacity=0.2,color=clr) ]) # P = plot3d(lambda x,y: H(x,y,ratio/10),(x,xx[0],xx[1]),(y,yy[0],yy[1]),opacity=0.1) # Now, accumulate all of the graphs into one grouped graph. P = sum(P_list[i] for i in range(len(P_list))) if show_3d: show(P,frame=true,axes=false,xmin=xx[0],xmax=xx[1],ymin=yy[0],ymax=yy[1],stereo='redcyan',figsize=(6,9),viewer='jmol',spin=dospin) else: show(P,frame=true,axes=false,xmin=xx[0],xmax=xx[1],ymin=yy[0],ymax=yy[1],figsize=(6,9),viewer='jmol',spin=dospin) }}} {{attachment:3D_Irregular_Volume.png}} == Lateral Surface Area (FIXME in Jupyter) == by John Travis http://sagenb.mc.edu/home/pub/89/ {{{#!sagecell ## Display and compute the area of the lateral surface between two surfaces ## corresponding to the (scalar) line integral ## John Travis ## Spring 2011 ## var('x,y,t,s') @interact(layout=dict(top=[['f','u'],['g','v']], left=[['a'],['b'],['in_3d'],['smoother']], bottom=[['xx','yy']])) def _(f=input_box(default=6-4*x^2-y^2*2/5,label='Top = $f(x,y) = $',width=30), g=input_box(default=-2+sin(x)+sin(y),label='Bottom = $g(x,y) = $',width=30), u=input_box(default=cos(t),label=' $ x = u(t) = $',width=20), v=input_box(default=2*sin(t),label=' $ y = v(t) = $',width=20), a=input_box(default=0,label='$a = $',width=10), b=input_box(default=3*pi/2,label='$b = $',width=10), xx = range_slider(-5, 5, 1, default=(-1,1), label='x view'), yy = range_slider(-5, 5, 1, default=(-2,2), label='y view'), in_3d = checkbox(default=true,label='3D'), smoother=checkbox(default=false), auto_update=true): ds = sqrt(derivative(u,t)^2+derivative(v,t)^2) # Set up the integrand to compute the line integral, making all attempts # to simplify the result so that it looks as nice as possible. A = (f(x=u,y=v)-g(x=u,y=v))*ds.simplify_trig().simplify() # It is not expected that Sage can actually perform the line integral calculation. # So, the result displayed may not be a numerical value as expected. # Creating a good but harder example that "works" is desirable. # If you want Sage to try, uncomment the lines below. # line_integral = integrate(A,t,a,b) # html(r'<align=center size=+1>Lateral Surface Area = $ %s $ </font>'%latex(line_integral)) line_integral_approx = numerical_integral(A,a,b)[0] html(r'<font align=center size=+1>Lateral Surface $ \approx $ %s</font>'%str(line_integral_approx)) # Plot the top function z = f(x,y) that is being integrated. G = plot3d(f,(x,xx[0],xx[1]),(y,yy[0],yy[1]),opacity=0.2) G += plot3d(g,(x,xx[0],xx[1]),(y,yy[0],yy[1]),opacity=0.2) # Add space curves on the surfaces "above" the domain curve (u(t),v(t)) G += parametric_plot3d([u,v,g(x=u,y=v)],(t,a,b),thickness=2,color='red') G += parametric_plot3d([u,v,f(x=u,y=v)],(t,a,b),thickness=2,color='red') k=0 if smoother: delw = 0.025 lat_thick = 3 else: delw = 0.10 lat_thick = 10 for w in (a,a+delw,..,b): G += parametric_plot3d([u(t=w),v(t=w),s*f(x=u(t=w),y=v(t=w))+(1-s)*g(x=u(t=w),y=v(t=w))],(s,0,1),thickness=lat_thick,color='yellow',opacity=0.9) if in_3d: show(G,stereo='redcyan',spin=true) else: show(G,perspective_depth=true,spin=true) }}} {{attachment:Lateral_Surface.png}} == Parametric surface example (FIXME in Jupyter) == |
| Line 1120: | Line 1469: |
| {{{ | {{{#!sagecell |
| Line 1134: | Line 1483: |
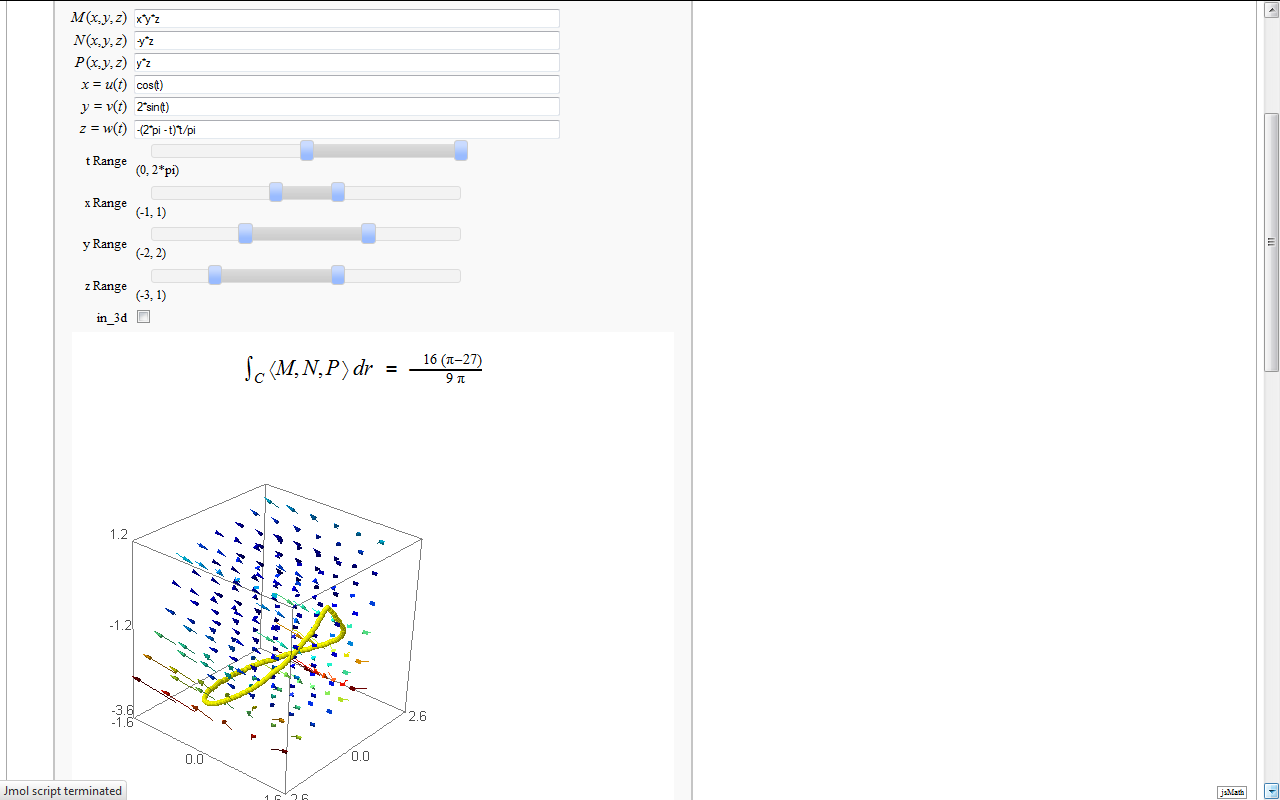
== Line Integrals in 3D Vector Field == by John Travis https://cloud.sagemath.com/projects/19575ea0-317e-402b-be57-368d04c113db/files/pub/2801-2901/2827-$%20%5Cint_%7BC%7D%20%5Cleft%20%5Clangle%20M,N,P%20%5Cright%20%5Crangle%20dr%20$%20=%20$%20%25s%20$.sagews {{{#!sagecell ## This worksheet interactively computes and displays the line integral of a 3D vector field ## over a given smooth curve C ## ## John Travis ## Mississippi College ## 06/16/11 ## ## An updated version of this worksheet may be available at http://sagenb.mc.edu ## var('x,y,z,t,s') @interact def _(M=input_box(default=x*y*z,label="$M(x,y,z)$"), N=input_box(default=-y*z,label="$N(x,y,z)$"), P=input_box(default=z*y,label="$P(x,y,z)$"), u=input_box(default=cos(t),label="$x=u(t)$"), v=input_box(default=2*sin(t),label="$y=v(t)$"), w=input_box(default=t*(t-2*pi)/pi,label="$z=w(t)$"), tt = range_slider(-2*pi, 2*pi, pi/6, default=(0,2*pi), label='t Range'), xx = range_slider(-5, 5, 1, default=(-1,1), label='x Range'), yy = range_slider(-5, 5, 1, default=(-2,2), label='y Range'), zz = range_slider(-5, 5, 1, default=(-3,1), label='z Range'), in_3d=checkbox(true)): # setup the parts and then compute the line integral dr = [derivative(u(t),t),derivative(v(t),t),derivative(w(t),t)] A = (M(x=u(t),y=v(t),z=w(t))*dr[0] +N(x=u(t),y=v(t),z=w(t))*dr[1] +P(x=u(t),y=v(t),z=w(t))*dr[2]) global line_integral line_integral = integral(A(t=t),t,tt[0],tt[1]) html(r'<h2 align=center>$ \int_{C} \left \langle M,N,P \right \rangle dr $ = $ %s $ </h2>'%latex(line_integral)) G = plot_vector_field3d((M,N,P),(x,xx[0],xx[1]),(y,yy[0],yy[1]),(z,zz[0],zz[1]),plot_points=6) G += parametric_plot3d([u,v,w],(t,tt[0],tt[1]),thickness='5',color='yellow') if in_3d: show(G,stereo='redcyan',spin=true) else: show(G,perspective_depth=true) }}} {{attachment:3D_Line_Integral.png}} |
Sage Interactions - Calculus
goto interact main page
Contents
-
Sage Interactions - Calculus
- Root Finding Using Bisection
- Newton's Method
- A contour map and 3d plot of two inverse distance functions
- A simple tangent line grapher
- Numerical integrals with the midpoint rule
- Numerical integrals with various rules
- Some polar parametric curves
- Function tool
- Newton-Raphson Root Finding
- Coordinate Transformations (FIXME in Jupyter)
- Taylor Series
- Illustration of the precise definition of a limit
- A graphical illustration of sin(x)/x -> 1 as x-> 0
- Quadric Surface Plotter
- The midpoint rule for numerically integrating a function of two variables
- Gaussian (Legendre) quadrature
- Vector Calculus, 2-D Motion
- Vector Calculus, 3-D Motion (FIXME)
- Multivariate Limits by Definition
- Directional Derivatives
- 3D graph with points and curves
- Approximating function in two variables by differential
- Taylor approximations in two variables
- Volumes over non-rectangular domains
- Lateral Surface Area (FIXME in Jupyter)
- Parametric surface example (FIXME in Jupyter)
- Line Integrals in 3D Vector Field
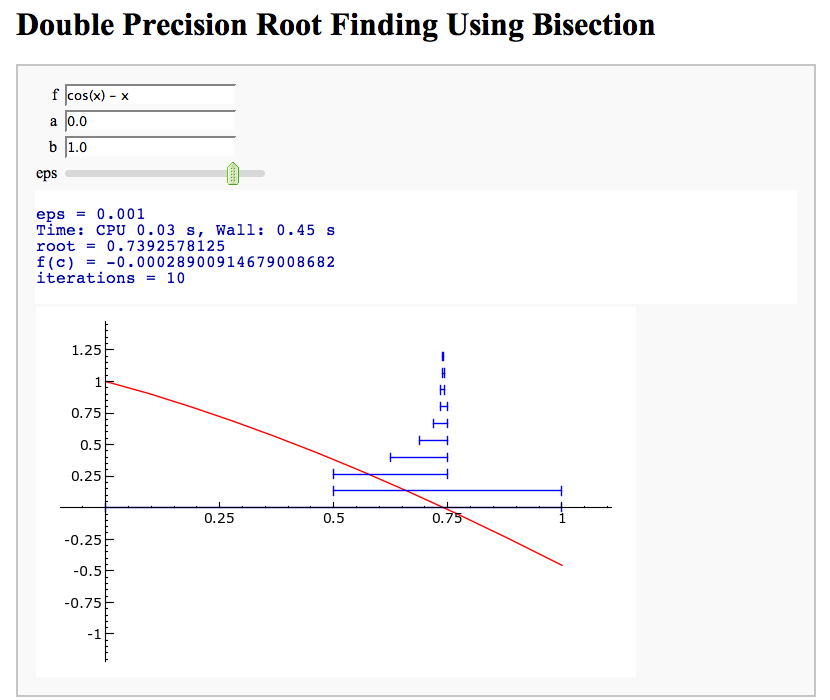
Root Finding Using Bisection
by William Stein
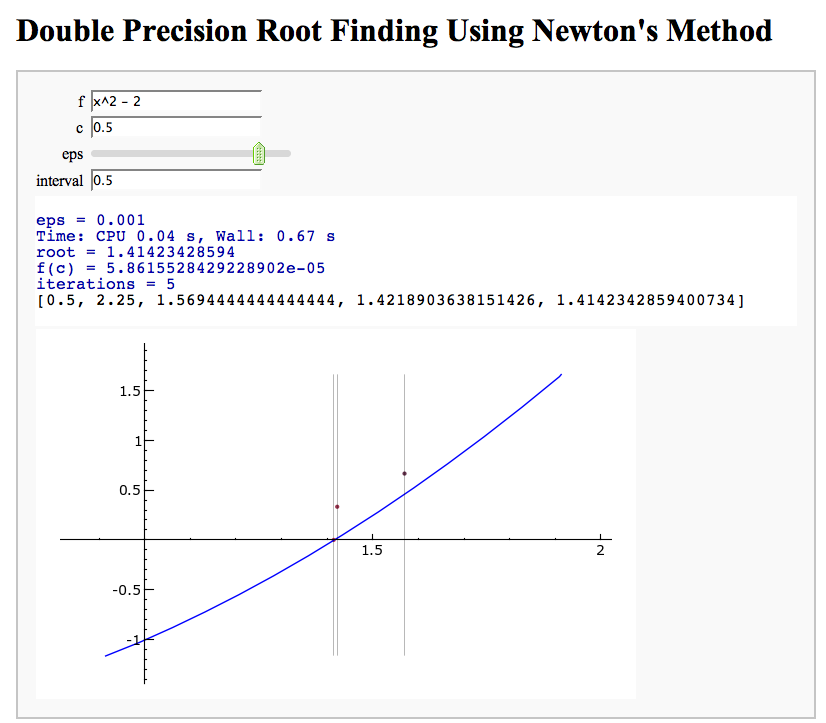
Newton's Method
Note that there is a more complicated Newton's method below.
by William Stein
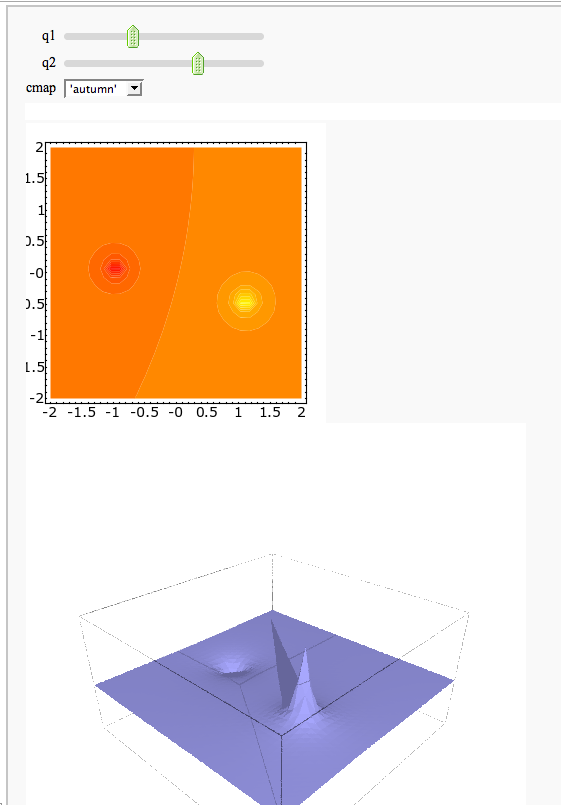
A contour map and 3d plot of two inverse distance functions
by William Stein
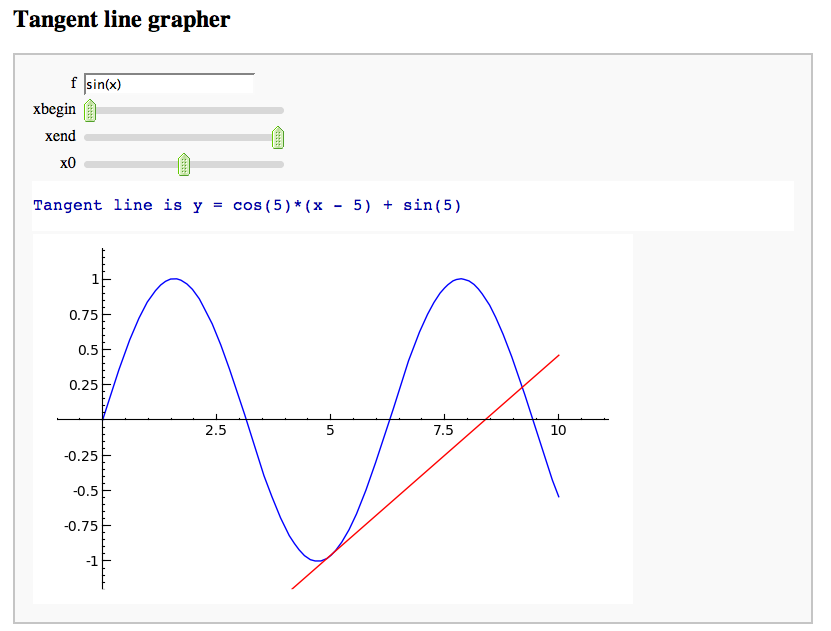
A simple tangent line grapher
by Marshall Hampton
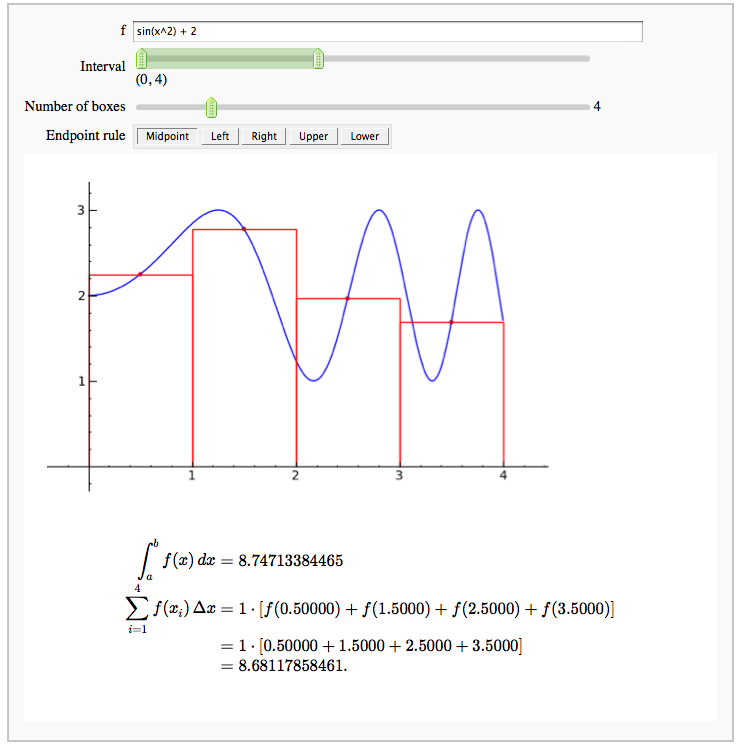
Numerical integrals with the midpoint rule
by Marshall Hampton

Numerical integrals with various rules
by Nick Alexander (based on the work of Marshall Hampton)
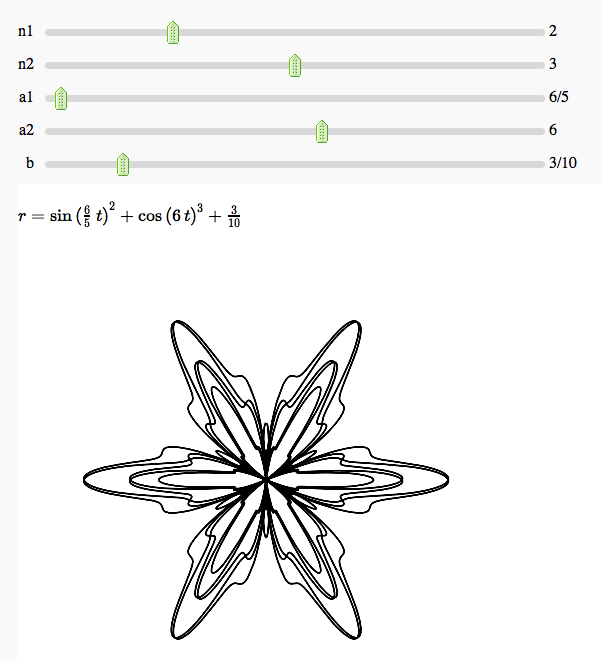
Some polar parametric curves
by Marshall Hampton. This is not very general, but could be modified to show other families of polar curves.
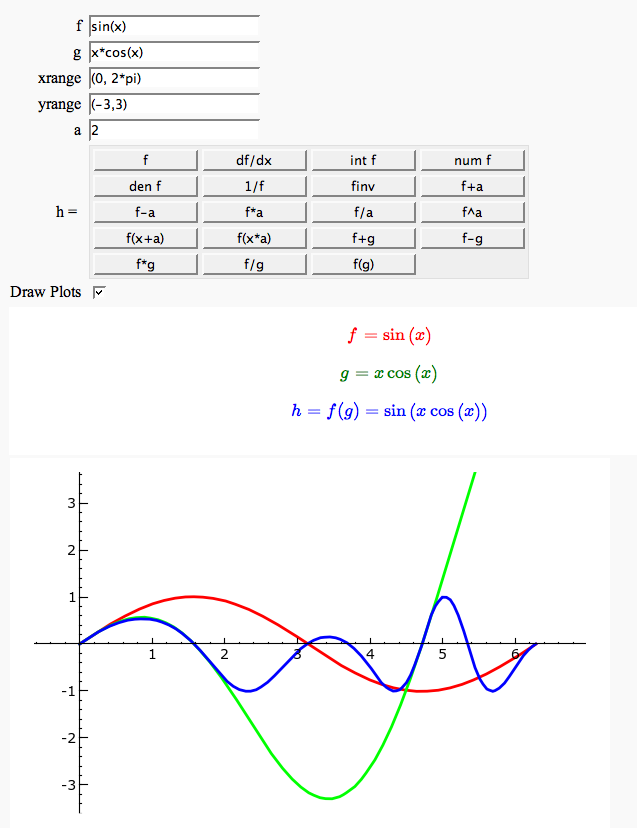
Function tool
Enter symbolic functions f, g, and a, a range, then click the appropriate button to compute and plot some combination of f, g, and a along with f and g. This is inspired by the Matlab funtool GUI.
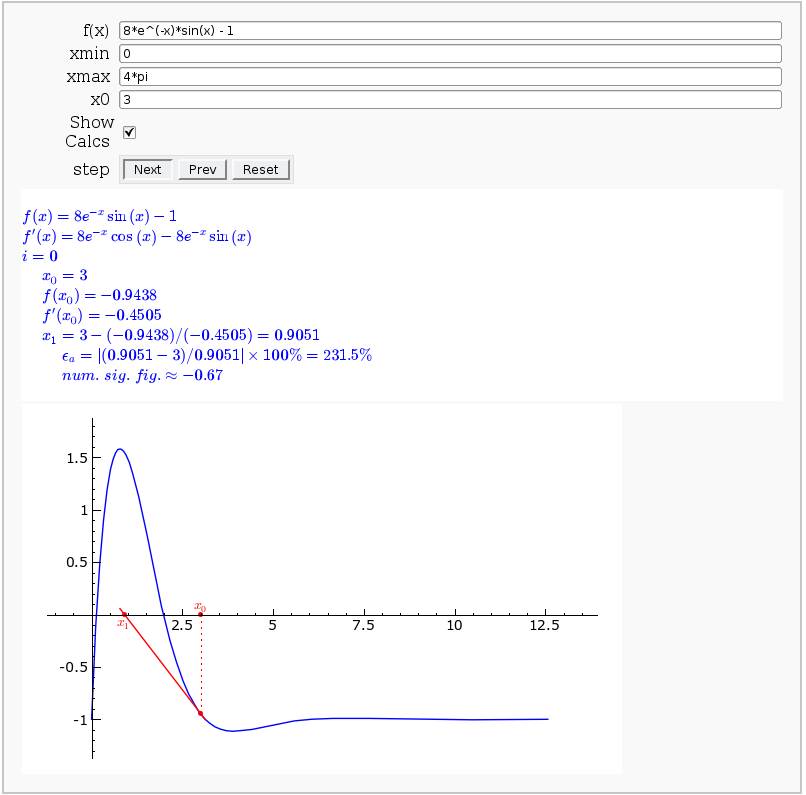
Newton-Raphson Root Finding
by Neal Holtz
This allows user to display the Newton-Raphson procedure one step at a time. It uses the heuristic that, if any of the values of the controls change, then the procedure should be re-started, else it should be continued.
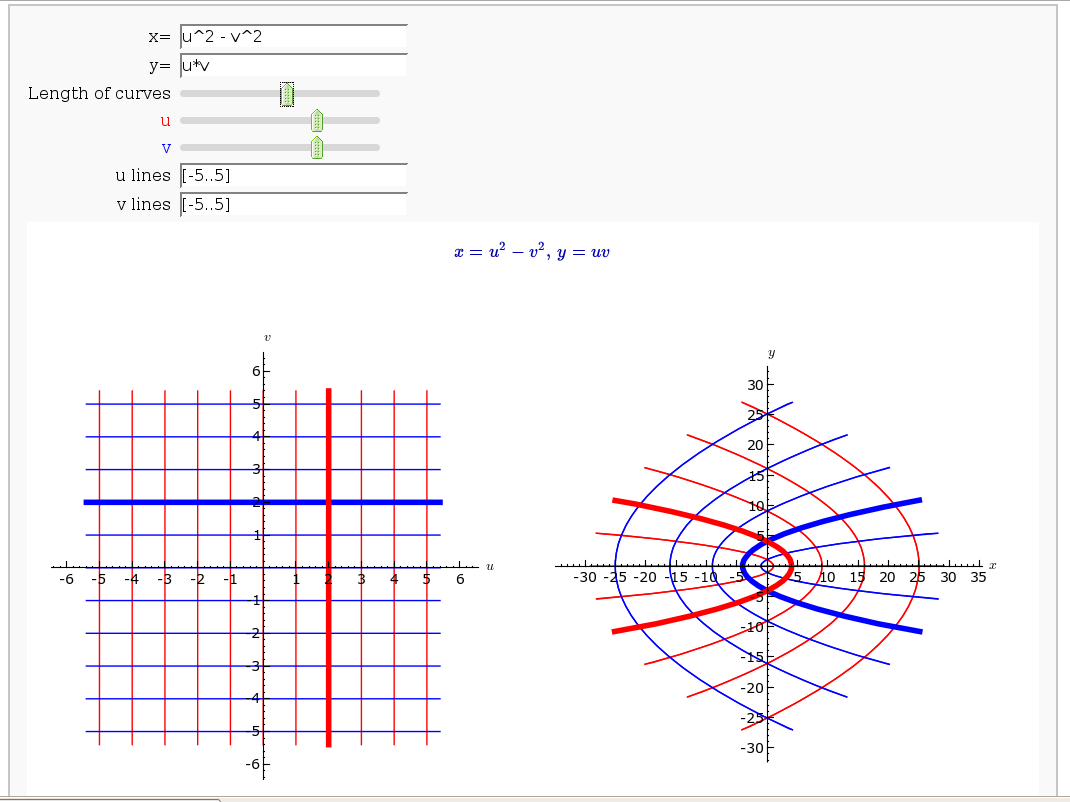
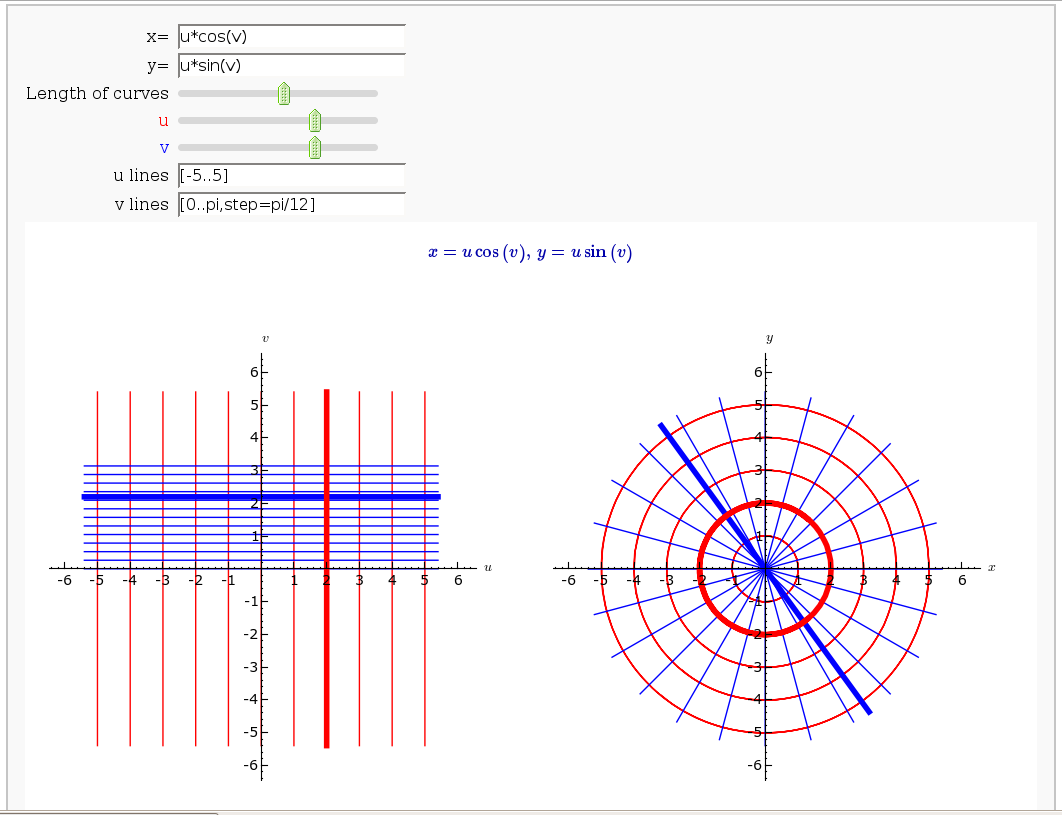
Coordinate Transformations (FIXME in Jupyter)
by Jason Grout
Taylor Series
by Harald Schilly
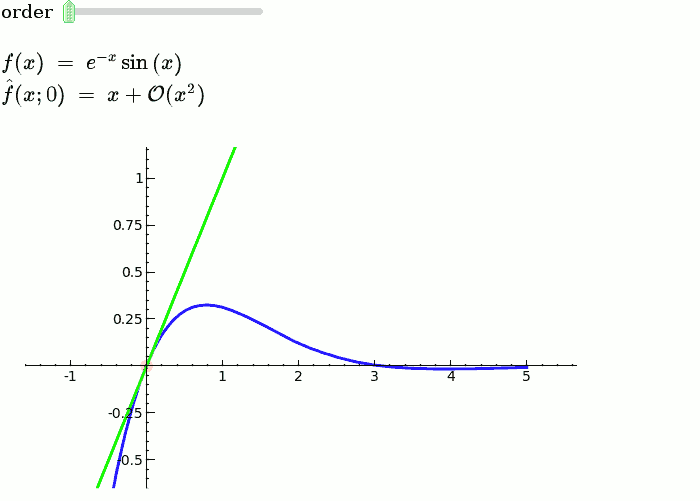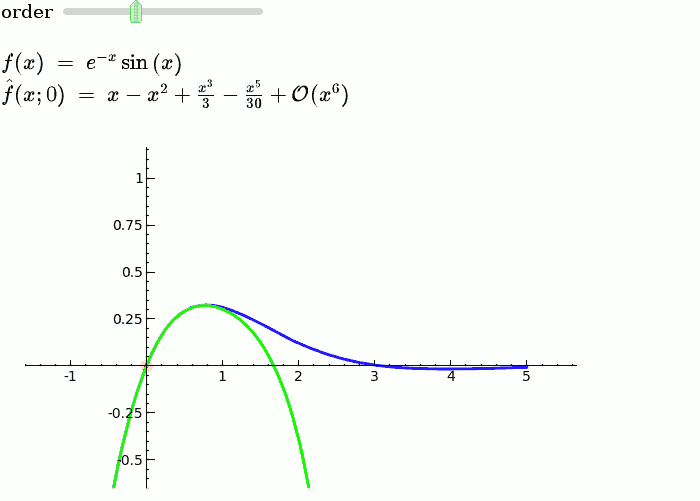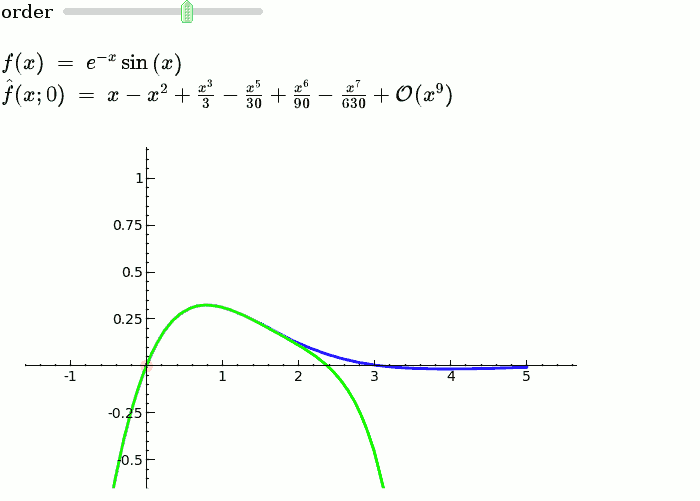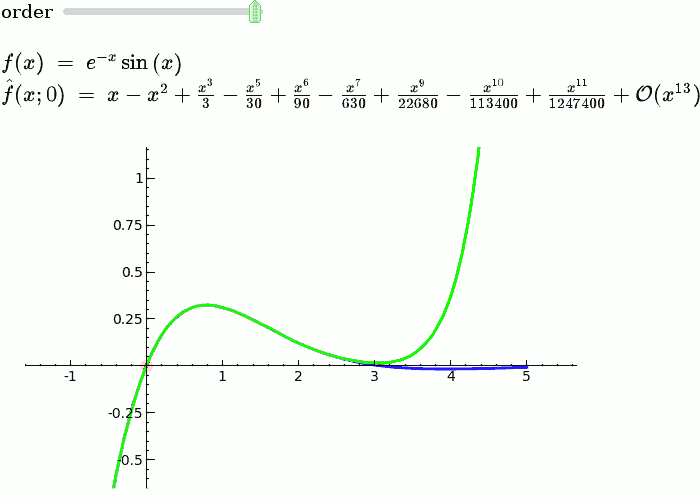
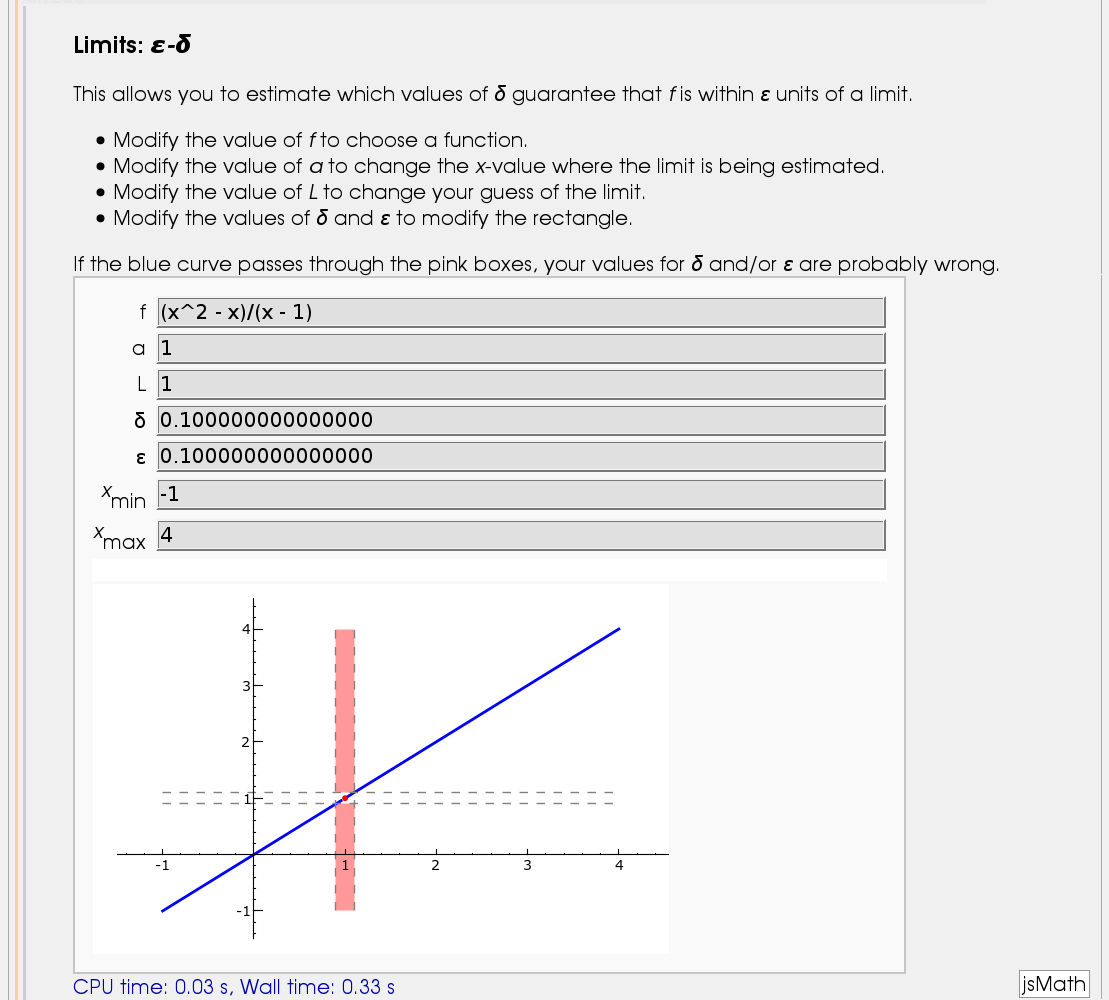
Illustration of the precise definition of a limit
by John Perry
I'll break tradition and put the image first. Apologies if this is Not A Good Thing.
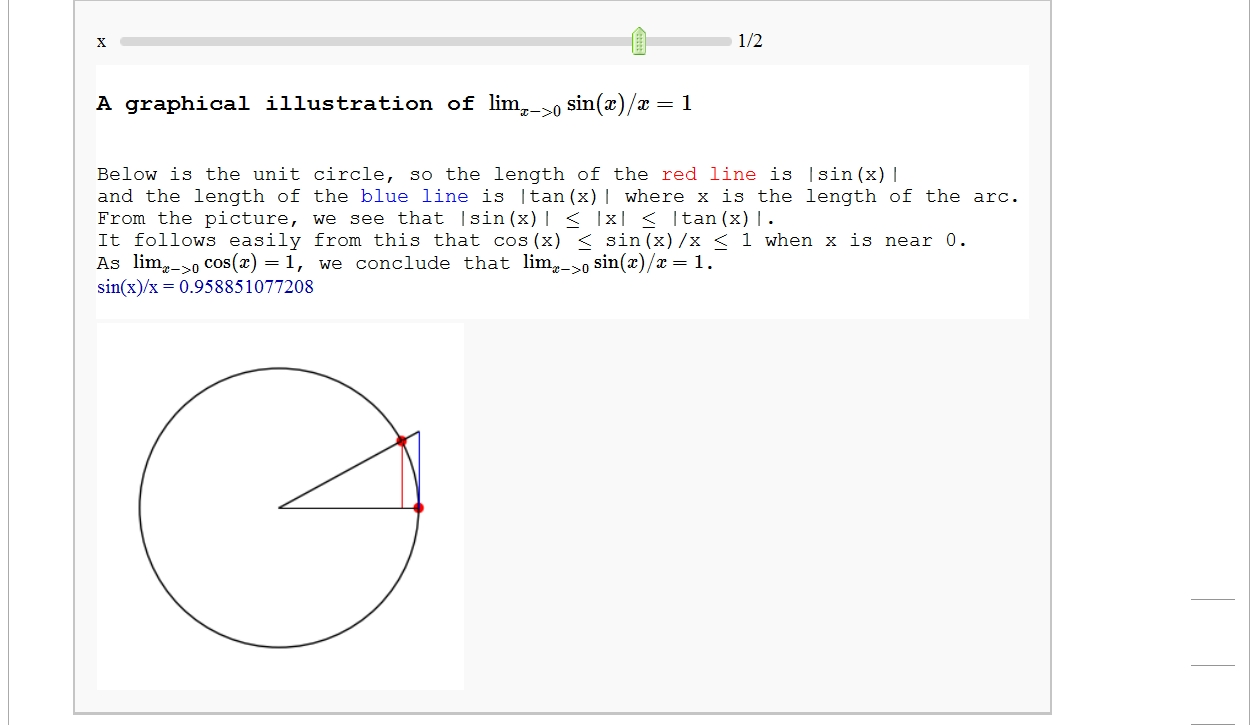
A graphical illustration of sin(x)/x -> 1 as x-> 0
by Wai Yan Pong
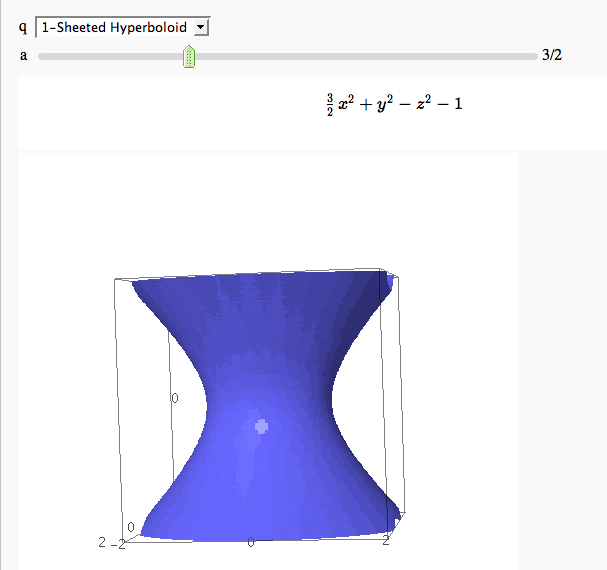
Quadric Surface Plotter
by Marshall Hampton. This is pretty simple, so I encourage people to spruce it up. In particular, it isn't set up to show all possible types of quadrics.
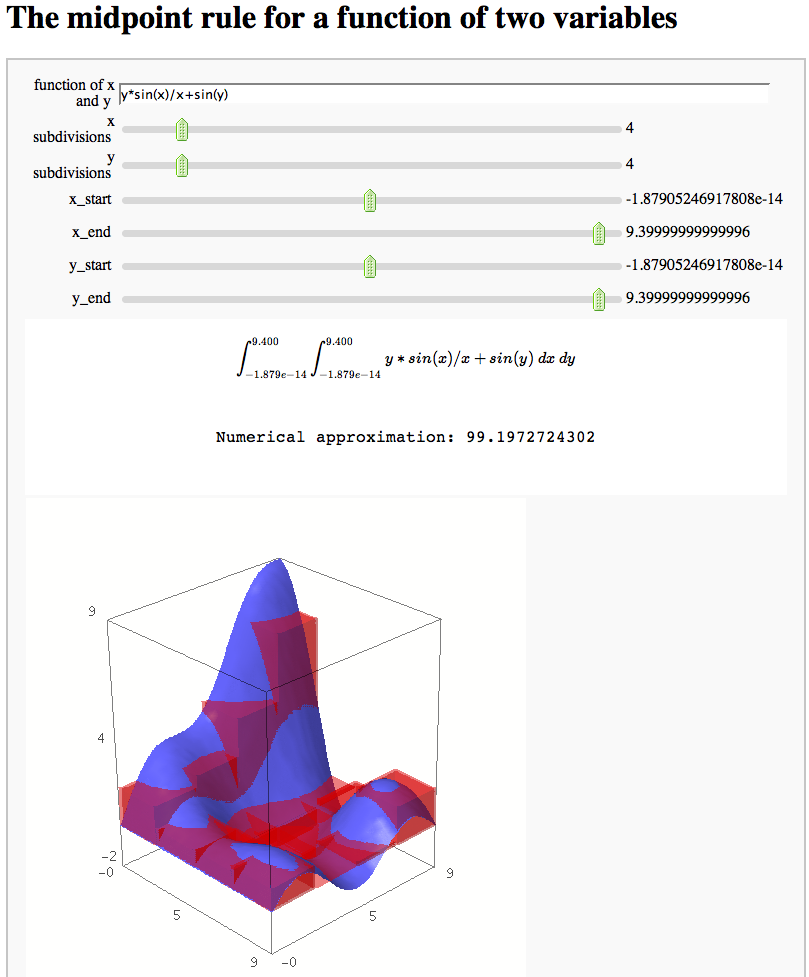
The midpoint rule for numerically integrating a function of two variables
by Marshall Hampton
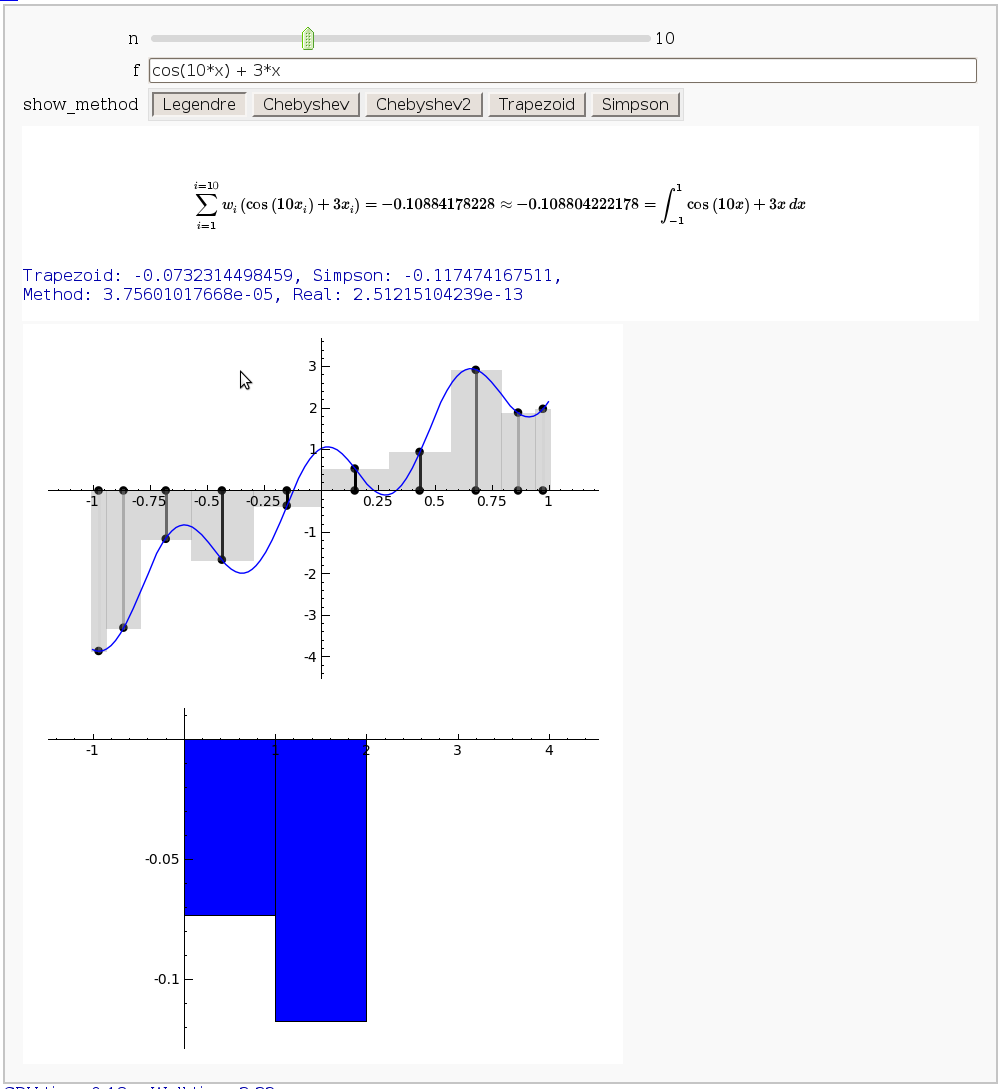
Gaussian (Legendre) quadrature
by Jason Grout
The output shows the points evaluated using Gaussian quadrature (using a weight of 1, so using Legendre polynomials). The vertical bars are shaded to represent the relative weights of the points (darker = more weight). The error in the trapezoid, Simpson, and quadrature methods is both printed out and compared through a bar graph. The "Real" error is the error returned from scipy on the definite integral.
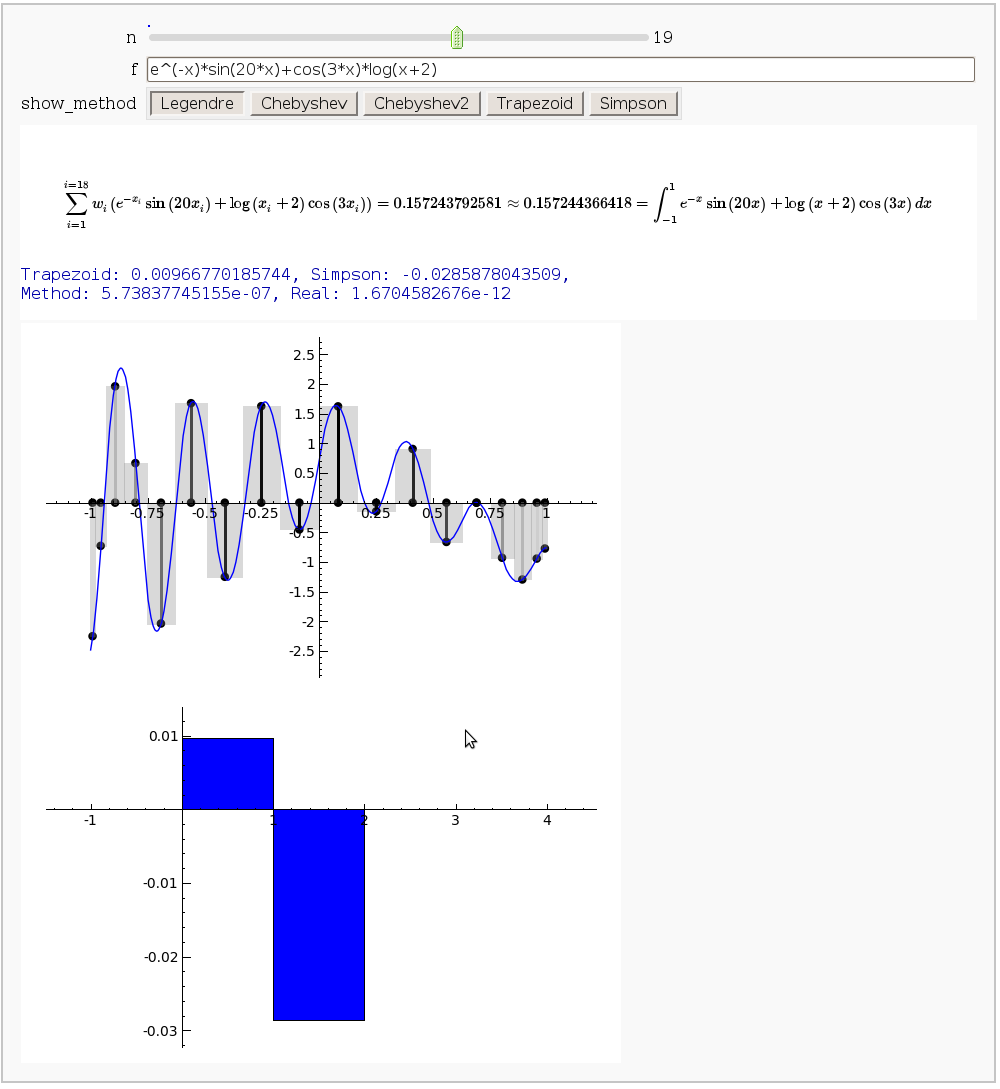
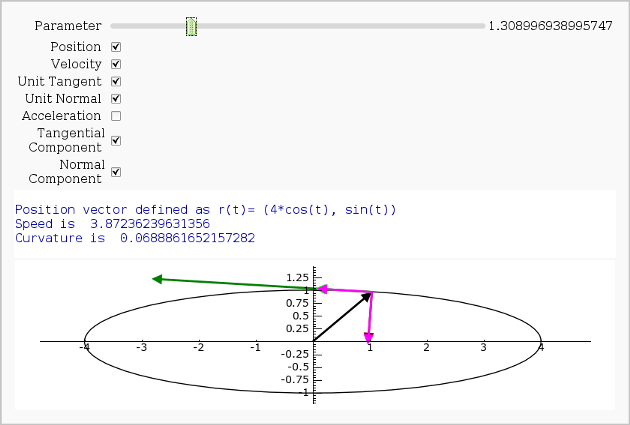
Vector Calculus, 2-D Motion
By Rob Beezer
A fast_float() version is available in a worksheet
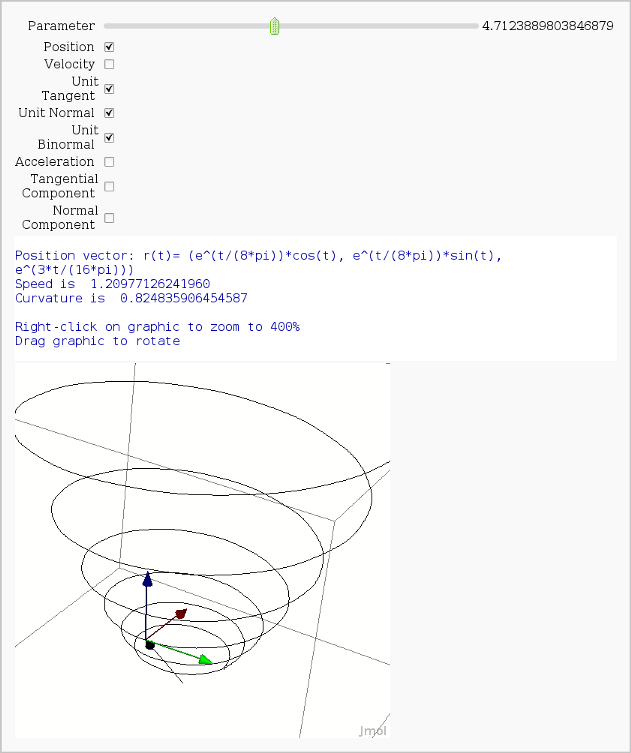
Vector Calculus, 3-D Motion (FIXME)
by Rob Beezer
Available as a worksheet
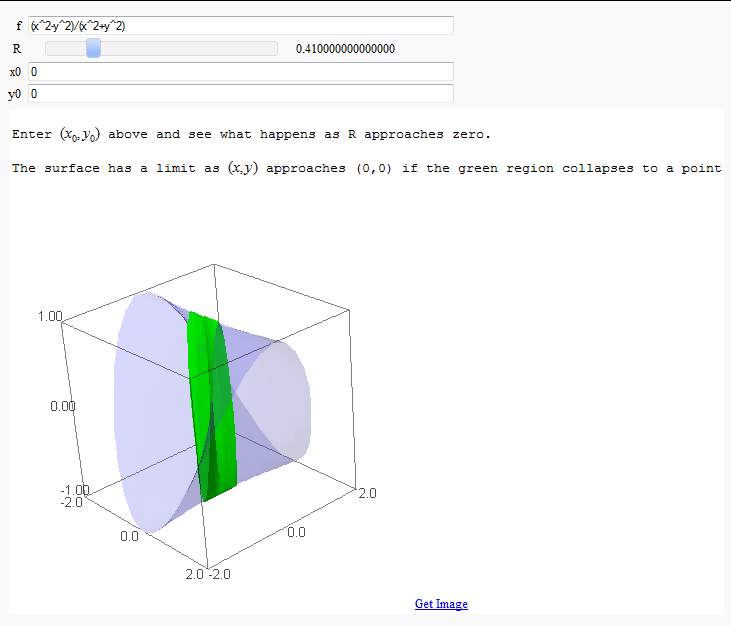
Multivariate Limits by Definition
by John Travis
http://sagenb.mc.edu/home/pub/97/
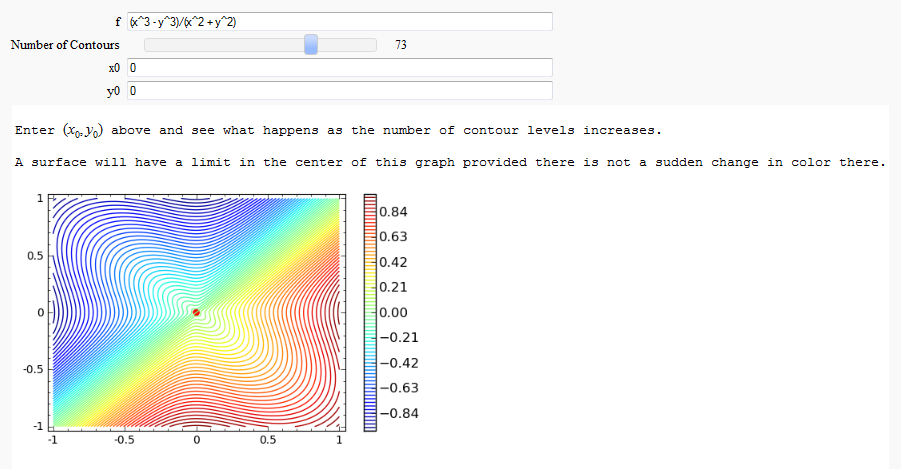
Directional Derivatives
This interact displays graphically a tangent line to a function, illustrating a directional derivative (the slope of the tangent line).
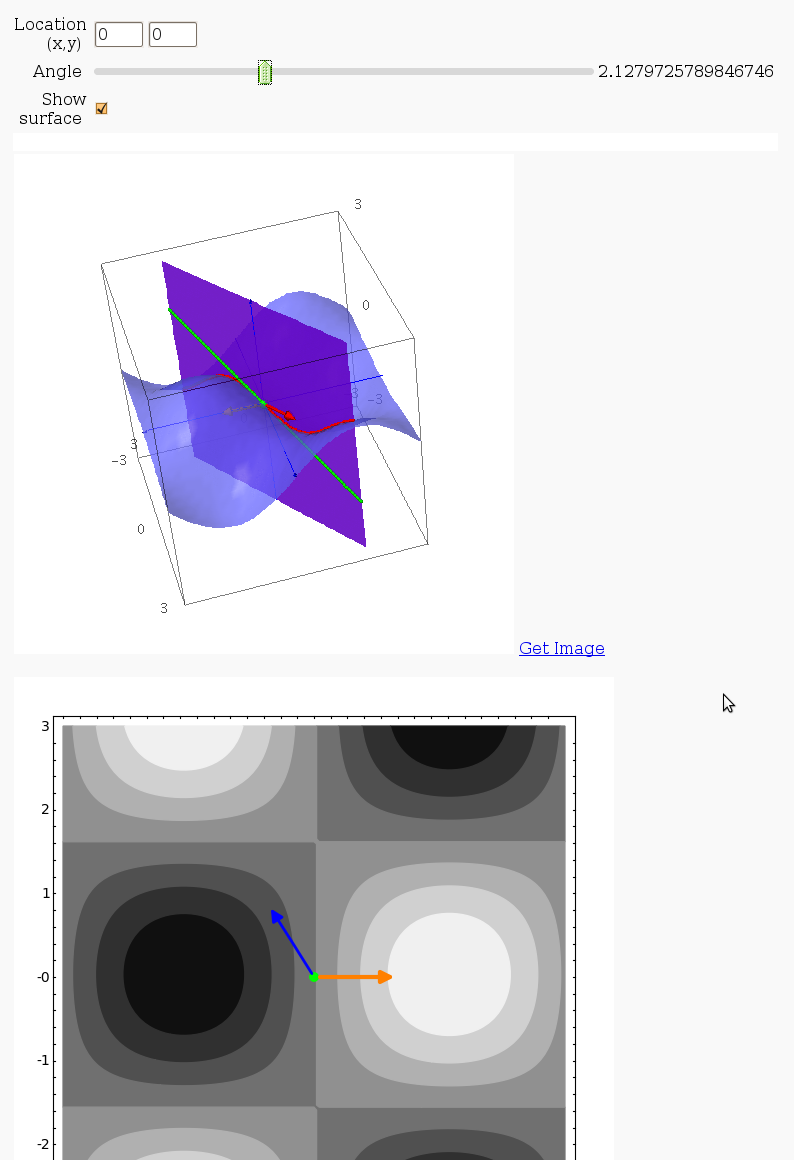
3D graph with points and curves
By Robert Marik
This sagelet is handy when showing local, constrained and absolute maxima and minima in two variables. Available as a worksheet
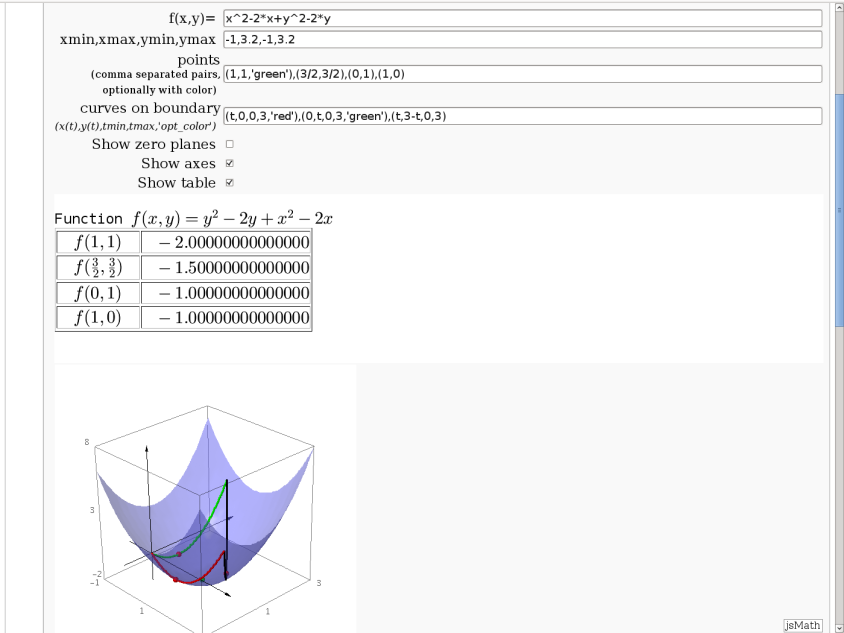
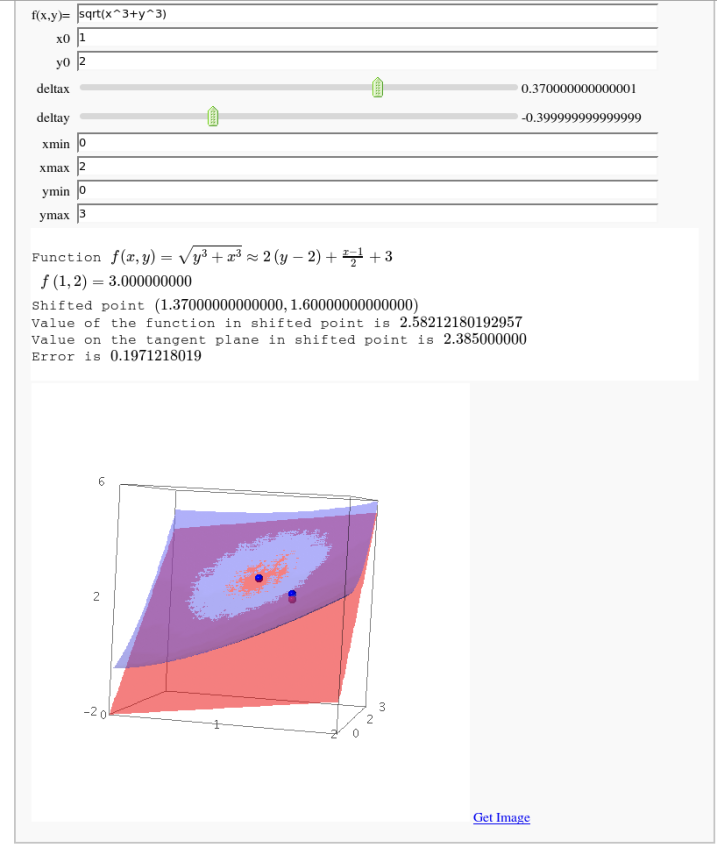
Approximating function in two variables by differential
by Robert Marik
Taylor approximations in two variables
by John Palmieri
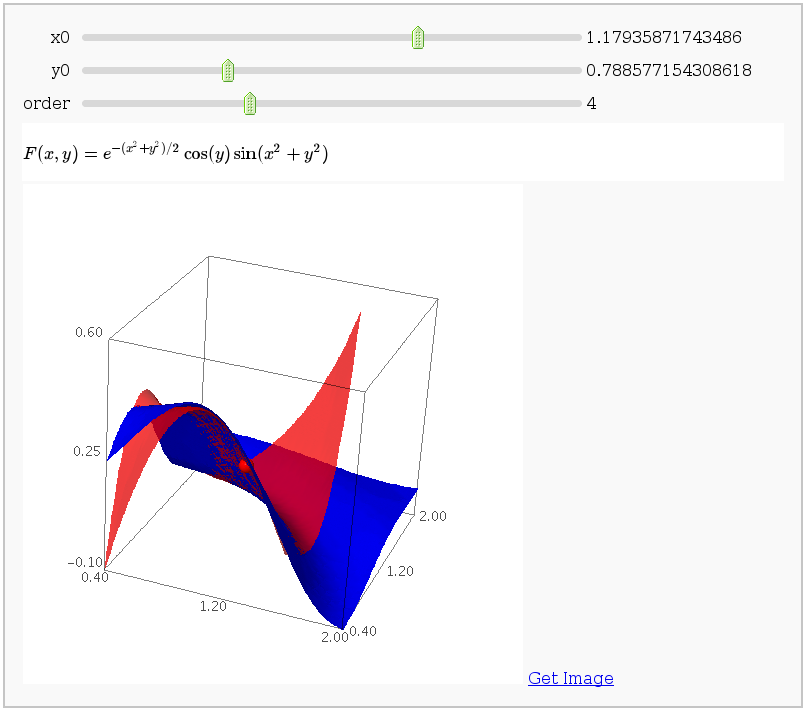
This displays the nth order Taylor approximation, for n from 1 to 10, of the function sin(x2 + y2) cos(y) exp(-(x2+y2)/2).
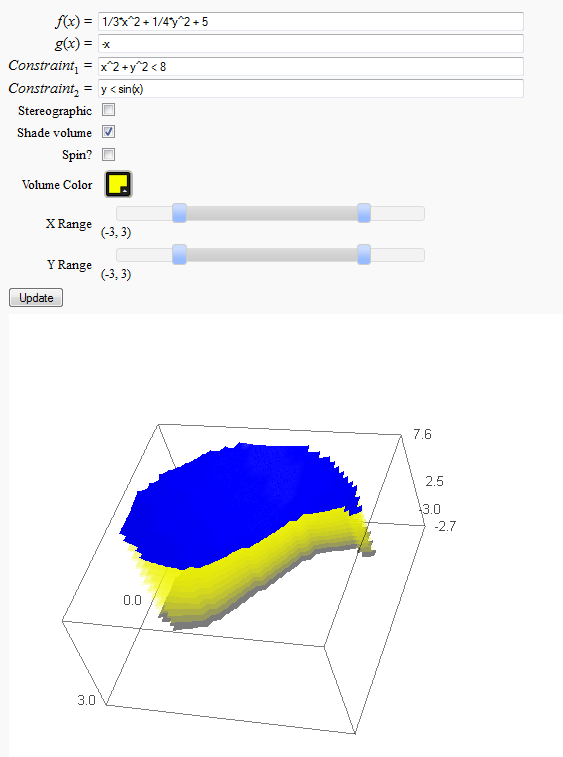
Volumes over non-rectangular domains
by John Travis
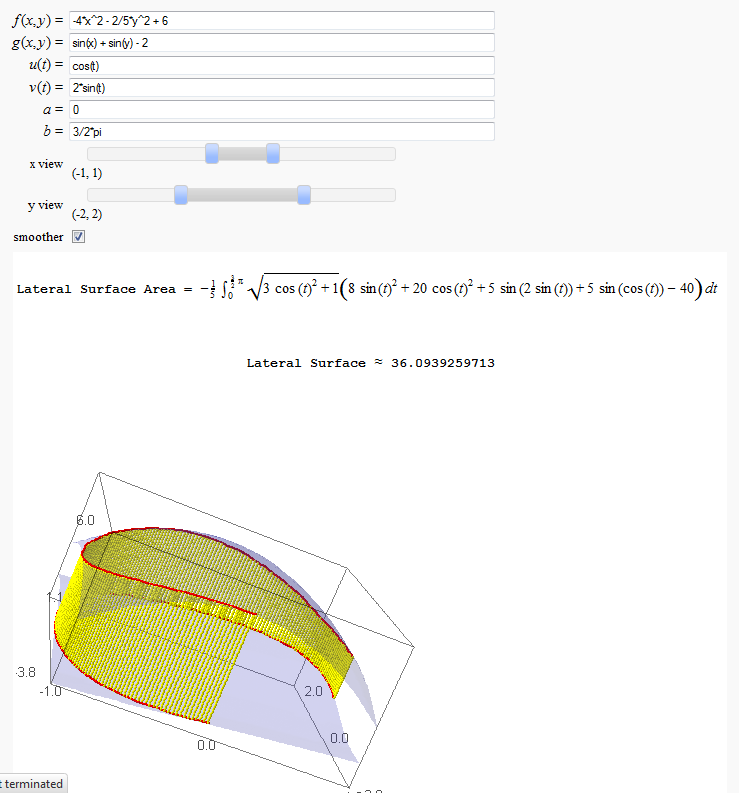
Lateral Surface Area (FIXME in Jupyter)
by John Travis
http://sagenb.mc.edu/home/pub/89/
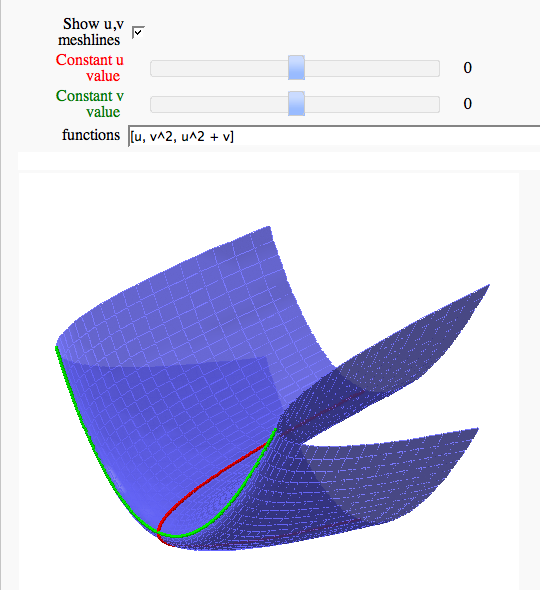
Parametric surface example (FIXME in Jupyter)
by Marshall Hampton
Line Integrals in 3D Vector Field
by John Travis