|
Size: 10742
Comment: py3 print
|
Size: 10824
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 6: | Line 6: |
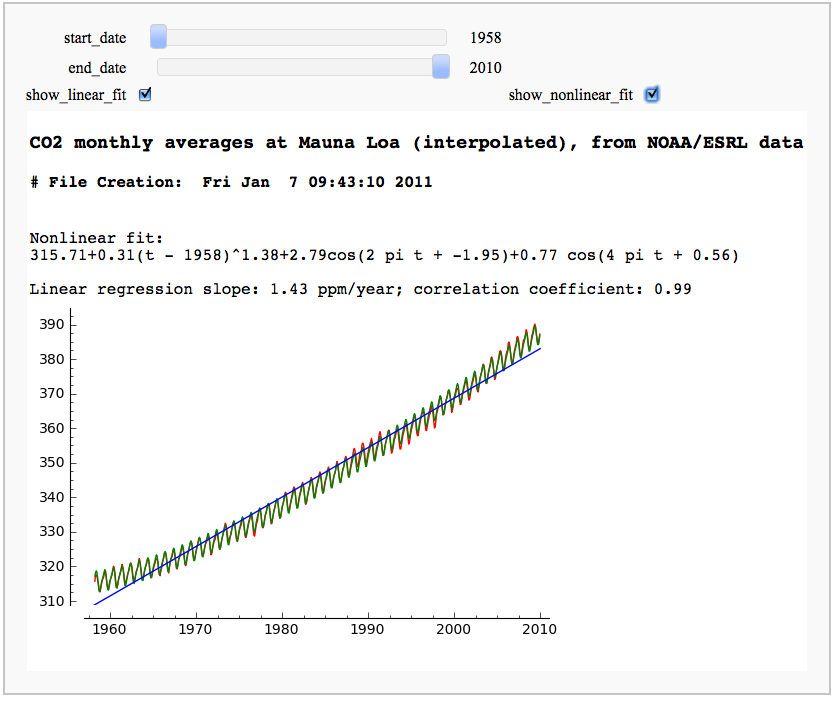
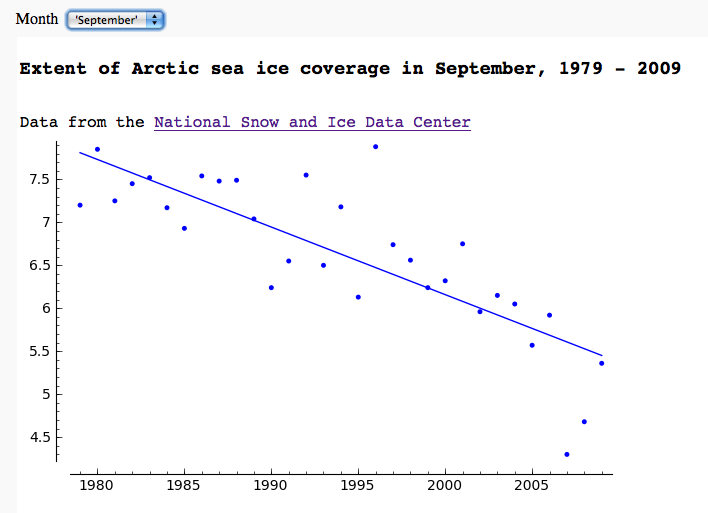
== CO2 data plot, fetched from NOAA == by Marshall Hampton One can do many things with scipy.stats. This only scratches the surface. {{{#!sagecell from scipy.optimize import leastsq import urllib.request as U import scipy.stats as Stat import time current_year = time.localtime().tm_year co2bytedata = U.urlopen('ftp://ftp.cmdl.noaa.gov/ccg/co2/trends/co2_mm_mlo.txt').readlines() co2data = [d.decode() for d in co2bytedata] datalines = [] for a_line in co2data: if a_line.find('Creation:') != -1: cdate = a_line if a_line[0] != '#': temp = a_line.replace('\n','').split(' ') temp = [float(q) for q in temp if q != ''] datalines.append(temp) npi = RDF(pi) @interact(layout=[['start_date'],['end_date'],['show_linear_fit','show_nonlinear_fit']]) def mauna_loa_co2(start_date = slider(1958,current_year,1,1958), end_date = slider(1958, current_year,1,current_year-1), show_linear_fit = checkbox(default=True), show_nonlinear_fit = checkbox(default=False)): htmls1 = '<h3>CO2 monthly averages at Mauna Loa (interpolated), from NOAA/ESRL data</h3>' htmls2 = '<h4>'+cdate+'</h4>' pretty_print(html(htmls1+htmls2)) sel_data = [[q[2],q[4]] for q in datalines if start_date <= q[2] <= end_date] outplot = list_plot(sel_data, plotjoined=True, rgbcolor=(1,0,0)) if show_nonlinear_fit: def powerlaw(t,a): return sel_data[0][1] + a[0]*(t-sel_data[0][0])^(a[1]) def res_fun(a): return [q[1]-powerlaw(q[0],a) for q in sel_data] def fitcos(t,a): return a[0]*cos(t*2*npi+a[1])+a[2]*cos(t*4*npi+a[3]) def res_fun2(a): return [q[1]-fitcos(q[0],a) for q in resids] a1 = leastsq(res_fun,[1/2.4,1.3])[0] resids = [[q[0],q[1] - powerlaw(q[0],a1)] for q in sel_data] a2 = leastsq(res_fun2, [3,0,1,0])[0] r2_plot = list_plot([[q[0],powerlaw(q[0],a1)+fitcos(q[0],a2)] for q in resids], rgbcolor='green',plotjoined=True) outplot = outplot + r2_plot var('t') formula1 = '%.2f+%.2f(t - %d)^%.2f'%(sel_data[0][1],a1[0],sel_data[0][0],a1[1]) formula2 = '%.2fcos(2 pi t + %.2f)+%.2f cos(4 pi t + %.2f)'%(a2[0],a2[1],a2[2],a2[3]) pretty_print(html('Nonlinear fit: <br>%s<br>'%(formula1+'+'+formula2))) if show_linear_fit: slope, intercept, r, ttprob, stderr = Stat.linregress(sel_data) outplot = outplot + plot(slope*x+intercept,start_date,end_date) pretty_print(html('Linear regression slope: %.2f ppm/year; correlation coefficient: %.2f'%(slope,r))) var('x,y') c_max = max([q[1] for q in sel_data]) c_min = min([q[1] for q in sel_data]) show(outplot, xmin = start_date, ymin = c_min-2, axes = True, xmax = end_date, ymax = c_max+3, frame = False) }}} {{attachment:co2c.png}} == Arctic sea ice extent data plot, fetched from NSIDC == by Marshall Hampton {{{#!sagecell import urllib2, csv months = ['Jan','Feb','Mar','Apr','May','Jun','Jul','Aug','Sep','Oct','Nov','Dec'] longmonths = ['January','February','March','April','May','June','July','August','September','October','November','December'] @interact def iceplotter(month = selector(zip(range(1,13),longmonths),default = (4, 'April'),label="Month")): month_str = months[month-1] + '/N_%02d_area.txt'%(month) dialect=csv.excel dialect.skipinitialspace = True icedata_f = urllib2.urlopen('ftp://sidads.colorado.edu/DATASETS/NOAA/G02135/%s'%month_str) cr = csv.reader(icedata_f,delimiter=' ', dialect=dialect) icedata = list(cr) icedata = [x for x in icedata[1:] if len(x)==6 and N(x[5])>0] lp = list_plot([[N(x[0]),N(x[4])] for x in icedata]) def lin_regress(xdata, ydata): xmean = N(mean(xdata)) ymean = N(mean(ydata)) xm = vector(RDF,[q-xmean for q in xdata]) ym = vector(RDF,[q-ymean for q in ydata]) xy = xm.inner_product(ym) xx = xm.inner_product(xm) slope = xy/xx intercept = ymean - slope*xmean return slope, intercept years = [N(x[0]) for x in icedata] ice = [N(x[4]) for x in icedata] slope, inter = lin_regress(years,ice) reg = plot(lambda x:slope*x+inter,(min(years),max(years))) html('<h3>Extent of Arctic sea ice coverage in %s, %d - %d</h3>'%(longmonths[month-1],min(years),max(years))) html('Data from the <a href="http://nsidc.org/">National Snow and Ice Data Center</a>') show(lp+reg, figsize = [7,4]) }}} {{attachment:seaice.png}} == Pie Chart from the Google Chart API == by Harald Schilly {{{#!sagecell # Google Chart API: http://code.google.com/apis/chart import urllib2 as inet from pylab import imshow @interact def gChart(title="Google Chart API plots Pie Charts!", color1=Color('purple'), color2=Color('black'), color3=Color('yellow'), val1=slider(0,1,.05,.5), val2=slider(0,1,.05,.3), val3=slider(0,1,.05,0.1), label=("Maths Physics Chemistry")): url = "http://chart.apis.google.com/chart?cht=p3&chs=600x300" url += '&chtt=%s&chts=000000,25'%title.replace(" ","+") url += '&chco=%s'%(','.join([color1.html_color()[1:],color2.html_color()[1:],color3.html_color()[1:]])) url += '&chl=%s'%label.replace(" ","|") url += '&chd=t:%s'%(','.join(map(str,[val1,val2,val3]))) print(url) html('<div style="border:3px dashed;text-align:center;padding:50px 0 50px 0"><img src="%s"></div>'%url) }}} {{attachment:interact_with_google_chart_api.png}} |
|
| Line 9: | Line 128: |
| (Need to fix plotting warnings as well as some stocks give index errors (like bsc, etc.) | (Need to fix plotting warnings as well as some stocks give index errors (like bcc), and Python 3 changes, etc.) |
| Line 121: | Line 240: |
| k = Y.keys(); k.sort() | k = list(Y); k.sort() |
| Line 129: | Line 248: |
== CO2 data plot, fetched from NOAA == by Marshall Hampton While support for R is rapidly improving, scipy.stats has a lot of useful stuff too. This only scratches the surface. {{{#!sagecell from scipy.optimize import leastsq import urllib2 as U import scipy.stats as Stat import time current_year = time.localtime().tm_year co2data = U.urlopen('ftp://ftp.cmdl.noaa.gov/ccg/co2/trends/co2_mm_mlo.txt').readlines() datalines = [] for a_line in co2data: if a_line.find('Creation:') != -1: cdate = a_line if a_line[0] != '#': temp = a_line.replace('\n','').split(' ') temp = [float(q) for q in temp if q != ''] datalines.append(temp) npi = RDF(pi) @interact(layout=[['start_date'],['end_date'],['show_linear_fit','show_nonlinear_fit']]) def mauna_loa_co2(start_date = slider(1958,current_year,1,1958), end_date = slider(1958, current_year,1,current_year-1), show_linear_fit = checkbox(default=True), show_nonlinear_fit = checkbox(default=False)): htmls1 = '<h3>CO2 monthly averages at Mauna Loa (interpolated), from NOAA/ESRL data</h3>' htmls2 = '<h4>'+cdate+'</h4>' html(htmls1+htmls2) sel_data = [[q[2],q[4]] for q in datalines if start_date <= q[2] <= end_date] outplot = list_plot(sel_data, plotjoined=True, rgbcolor=(1,0,0)) if show_nonlinear_fit: def powerlaw(t,a): return sel_data[0][1] + a[0]*(t-sel_data[0][0])^(a[1]) def res_fun(a): return [q[1]-powerlaw(q[0],a) for q in sel_data] def fitcos(t,a): return a[0]*cos(t*2*npi+a[1])+a[2]*cos(t*4*npi+a[3]) def res_fun2(a): return [q[1]-fitcos(q[0],a) for q in resids] a1 = leastsq(res_fun,[1/2.4,1.3])[0] resids = [[q[0],q[1] - powerlaw(q[0],a1)] for q in sel_data] a2 = leastsq(res_fun2, [3,0,1,0])[0] r2_plot = list_plot([[q[0],powerlaw(q[0],a1)+fitcos(q[0],a2)] for q in resids], rgbcolor='green',plotjoined=True) outplot = outplot + r2_plot var('t') formula1 = '%.2f+%.2f(t - %d)^%.2f'%(sel_data[0][1],a1[0],sel_data[0][0],a1[1]) formula2 = '%.2fcos(2 pi t + %.2f)+%.2f cos(4 pi t + %.2f)'%(a2[0],a2[1],a2[2],a2[3]) html('Nonlinear fit: <br>%s<br>'%(formula1+'+'+formula2)) if show_linear_fit: slope, intercept, r, ttprob, stderr = Stat.linregress(sel_data) outplot = outplot + plot(slope*x+intercept,start_date,end_date) html('Linear regression slope: %.2f ppm/year; correlation coefficient: %.2f'%(slope,r)) var('x,y') c_max = max([q[1] for q in sel_data]) c_min = min([q[1] for q in sel_data]) show(outplot, xmin = start_date, ymin = c_min-2, axes = True, xmax = end_date, ymax = c_max+3, frame = False) }}} {{attachment:co2c.png}} == Arctic sea ice extent data plot, fetched from NSIDC == by Marshall Hampton {{{#!sagecell import urllib2, csv months = ['Jan','Feb','Mar','Apr','May','Jun','Jul','Aug','Sep','Oct','Nov','Dec'] longmonths = ['January','February','March','April','May','June','July','August','September','October','November','December'] @interact def iceplotter(month = selector(zip(range(1,13),longmonths),default = (4, 'April'),label="Month")): month_str = months[month-1] + '/N_%02d_area.txt'%(month) dialect=csv.excel dialect.skipinitialspace = True icedata_f = urllib2.urlopen('ftp://sidads.colorado.edu/DATASETS/NOAA/G02135/%s'%month_str) cr = csv.reader(icedata_f,delimiter=' ', dialect=dialect) icedata = list(cr) icedata = [x for x in icedata[1:] if len(x)==6 and N(x[5])>0] lp = list_plot([[N(x[0]),N(x[4])] for x in icedata]) def lin_regress(xdata, ydata): xmean = N(mean(xdata)) ymean = N(mean(ydata)) xm = vector(RDF,[q-xmean for q in xdata]) ym = vector(RDF,[q-ymean for q in ydata]) xy = xm.inner_product(ym) xx = xm.inner_product(xm) slope = xy/xx intercept = ymean - slope*xmean return slope, intercept years = [N(x[0]) for x in icedata] ice = [N(x[4]) for x in icedata] slope, inter = lin_regress(years,ice) reg = plot(lambda x:slope*x+inter,(min(years),max(years))) html('<h3>Extent of Arctic sea ice coverage in %s, %d - %d</h3>'%(longmonths[month-1],min(years),max(years))) html('Data from the <a href="http://nsidc.org/">National Snow and Ice Data Center</a>') show(lp+reg, figsize = [7,4]) }}} {{attachment:seaice.png}} == Pie Chart from the Google Chart API == by Harald Schilly {{{#!sagecell # Google Chart API: http://code.google.com/apis/chart import urllib2 as inet from pylab import imshow @interact def gChart(title="Google Chart API plots Pie Charts!", color1=Color('purple'), color2=Color('black'), color3=Color('yellow'), val1=slider(0,1,.05,.5), val2=slider(0,1,.05,.3), val3=slider(0,1,.05,0.1), label=("Maths Physics Chemistry")): url = "http://chart.apis.google.com/chart?cht=p3&chs=600x300" url += '&chtt=%s&chts=000000,25'%title.replace(" ","+") url += '&chco=%s'%(','.join([color1.html_color()[1:],color2.html_color()[1:],color3.html_color()[1:]])) url += '&chl=%s'%label.replace(" ","|") url += '&chd=t:%s'%(','.join(map(str,[val1,val2,val3]))) print(url) html('<div style="border:3px dashed;text-align:center;padding:50px 0 50px 0"><img src="%s"></div>'%url) }}} {{attachment:interact_with_google_chart_api.png}} |
Sage Interactions - Web applications
goto interact main page
Contents
CO2 data plot, fetched from NOAA
by Marshall Hampton
One can do many things with scipy.stats. This only scratches the surface.
Arctic sea ice extent data plot, fetched from NSIDC
by Marshall Hampton
Pie Chart from the Google Chart API
by Harald Schilly

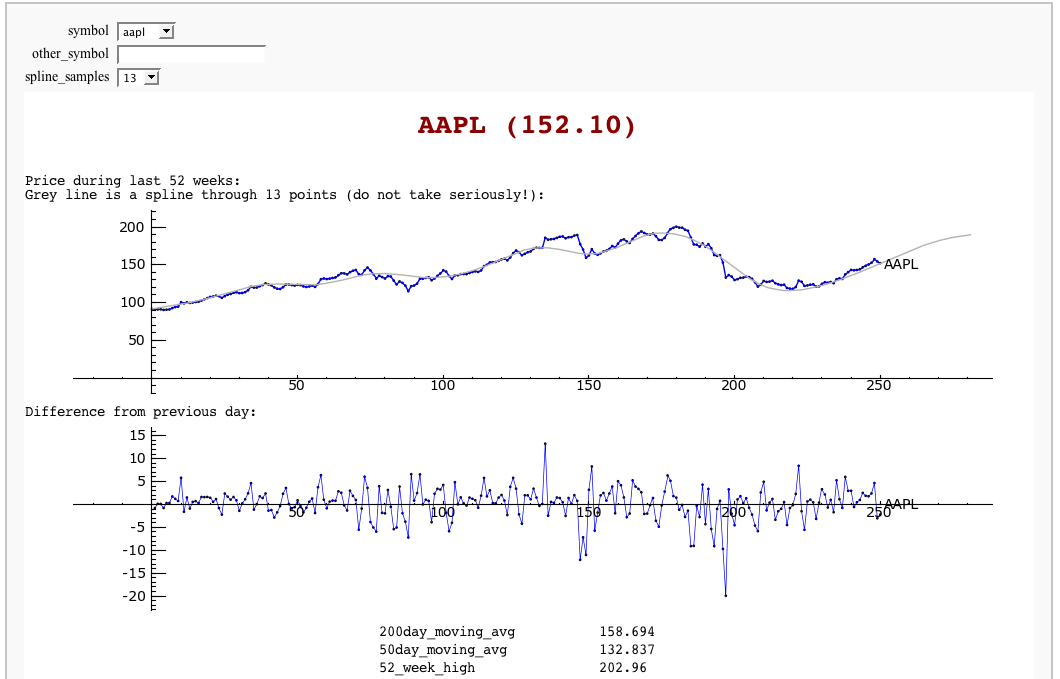
Stock Market data, fetched from Yahoo and Google FIXME
by William Stein
(Need to fix plotting warnings as well as some stocks give index errors (like bcc), and Python 3 changes, etc.)